Pharmacological interventions enhance virus-free generation of TRAC-replaced CAR T cells
- PMID: 35573047
- PMCID: PMC9062427
- DOI: 10.1016/j.omtm.2022.03.018
Pharmacological interventions enhance virus-free generation of TRAC-replaced CAR T cells
Abstract
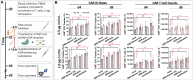
Chimeric antigen receptor (CAR) redirected T cells are potent therapeutic options against hematological malignancies. The current dominant manufacturing approach for CAR T cells depends on retroviral transduction. With the advent of gene editing, insertion of a CD19-CAR into the T cell receptor (TCR) alpha constant (TRAC) locus using adeno-associated viruses for gene transfer was demonstrated, and these CD19-CAR T cells showed improved functionality over their retrovirally transduced counterparts. However, clinical-grade production of viruses is complex and associated with extensive costs. Here, we optimized a virus-free genome-editing method for efficient CAR insertion into the TRAC locus of primary human T cells via nuclease-assisted homology-directed repair (HDR) using CRISPR-Cas and double-stranded template DNA (dsDNA). We evaluated DNA-sensor inhibition and HDR enhancement as two pharmacological interventions to improve cell viability and relative CAR knockin rates, respectively. While the toxicity of transfected dsDNA was not fully prevented, the combination of both interventions significantly increased CAR knockin rates and CAR T cell yield. Resulting TRAC-replaced CD19-CAR T cells showed antigen-specific cytotoxicity and cytokine production in vitro and slowed leukemia progression in a xenograft mouse model. Amplicon sequencing did not reveal significant indel formation at potential off-target sites with or without exposure to DNA-repair-modulating small molecules. With TRAC-integrated CAR+ T cell frequencies exceeding 50%, this study opens new perspectives to exploit pharmacological interventions to improve non-viral gene editing in T cells.
Keywords: CAR T cells; CRISPR-Cas9; HDR; TRAC; adoptive T cell therapy; chimeric antigen receptor; gene editing; knockin; non-viral cell manufacturing.
© 2022 The Author(s).
Conflict of interest statement
As part of a collaboration agreement between Charité Universitätsmedizin Berlin and Integrated DNA Technologies (IDT), IDT provided certain reagents (HDR enhancer v.2 and TRAC sgRNA used in some experiments) and performed GUIDE-seq analysis, HDR-enhancing small-molecule screen in Jurkat cells, and targeted sequencing of potential off-target sites. R.T., B.T., M.L.S., G.L.K., and A.M.J. are employees of IDT, which offers reagents for sale similar to some of the compounds described in the manuscript. Products and tools supplied by IDT are for research use only and not intended for diagnostic or therapeutic purposes. Purchaser and/or user are solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations. Lonza GmbH provided 96-well 4D-Nucleofector unit and some nucleofection reagents. A.W. and L. Akyüz are part-time employees of CheckImmune GmbH. A.R. and U.E.H. filed a patent application WO 2017211900A1 “Chimeric antigen receptor and CAR T cells that bind BCMA” related to the work with the BCMA-CAR disclosed in this paper. A.R. and U.E.H. have received research funding from Fate Therapeutics for work unrelated to the data generated in the manuscript.
Figures







References
-
- Schuster S.J., Bishop M.R., Tam C.S., Waller E.K., Borchmann P., McGuirk J.P., Jäger U., Jaglowski S., Andreadis C., Westin J.R., et al. Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. N. Engl. J. Med. 2019;380:45–56. - PubMed
-
- Brudno J.N., Maric I., Hartman S.D., Rose J.J., Wang M., Lam N., Stetler-Stevenson M., Salem D., Yuan C., Pavletic S., et al. T cells genetically modified to express an anti–B-cell maturation antigen chimeric antigen receptor cause remissions of poor-prognosis relapsed multiple myeloma. J. Clin. Oncol. 2018;36:2267–2280. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

