Studying Metabolism by NMR-Based Metabolomics
- PMID: 35573745
- PMCID: PMC9094115
- DOI: 10.3389/fmolb.2022.882487
Studying Metabolism by NMR-Based Metabolomics
Abstract
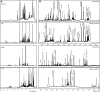
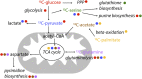
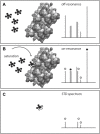
During the past few decades, the direct analysis of metabolic intermediates in biological samples has greatly improved the understanding of metabolic processes. The most used technologies for these advances have been mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy. NMR is traditionally used to elucidate molecular structures and has now been extended to the analysis of complex mixtures, as biological samples: NMR-based metabolomics. There are however other areas of small molecule biochemistry for which NMR is equally powerful. These include the quantification of metabolites (qNMR); the use of stable isotope tracers to determine the metabolic fate of drugs or nutrients, unravelling of new metabolic pathways, and flux through pathways; and metabolite-protein interactions for understanding metabolic regulation and pharmacological effects. Computational tools and resources for automating analysis of spectra and extracting meaningful biochemical information has developed in tandem and contributes to a more detailed understanding of systems biochemistry. In this review, we highlight the contribution of NMR in small molecule biochemistry, specifically in metabolic studies by reviewing the state-of-the-art methodologies of NMR spectroscopy and future directions.
Keywords: NMR; metabolism; metabolite-protein interactions; metabolomics; qNMR; stable isotopes.
Copyright © 2022 Moco.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Anklin C. (2016). “Chapter 3. Small-Volume NMR: Microprobes and Cryoprobes,” in Modern NMR Approaches to the Structure Elucidation of Natural Products, Instrumentation and Software. Editors Martin G. E., Williams A. J., Rovnyak D. (Cambridge, United Kingdom: Royal Society of Chemistry; ), 38–57. 10.1039/9781849735186-00038 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

