An antibody-escape estimator for mutations to the SARS-CoV-2 receptor-binding domain
- PMID: 35573973
- PMCID: PMC9092643
- DOI: 10.1093/ve/veac021
An antibody-escape estimator for mutations to the SARS-CoV-2 receptor-binding domain
Abstract
A key goal of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) surveillance is to rapidly identify viral variants with mutations that reduce neutralization by polyclonal antibodies elicited by vaccination or infection. Unfortunately, direct experimental characterization of new viral variants lags their sequence-based identification. Here we help address this challenge by aggregating deep mutational scanning data into an 'escape estimator' that estimates the antigenic effects of arbitrary combinations of mutations to the virus's spike receptor-binding domain. The estimator can be used to intuitively visualize how mutations impact polyclonal antibody recognition and score the expected antigenic effect of combinations of mutations. These scores correlate with neutralization assays performed on SARS-CoV-2 variants and emphasize the ominous antigenic properties of the recently described Omicron variant. An interactive version of the estimator is at https://jbloomlab.github.io/SARS2_RBD_Ab_escape_maps/escape-calc/ (last accessed 11 March 2022), and we provide a Python module for batch processing. Currently the calculator uses primarily data for antibodies elicited by Wuhan-Hu-1-like vaccination or infection and so is expected to work best for calculating escape from such immunity for mutations relative to early SARS-CoV-2 strains.
Keywords: Omicron; SARS-CoV-2 variants; antibody escape; deep mutational scanning; epitope.
© The Author(s) 2022. Published by Oxford University Press.
Figures
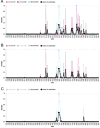

Update of
-
An antibody-escape calculator for mutations to the SARS-CoV-2 receptor-binding domain.bioRxiv [Preprint]. 2021 Dec 7:2021.12.04.471236. doi: 10.1101/2021.12.04.471236. bioRxiv. 2021. Update in: Virus Evol. 2022 May 11;8(1):veac021. doi: 10.1093/ve/veac021. PMID: 34909770 Free PMC article. Updated. Preprint.
References
-
- de Oliveira T. (2021) <https://twitter.com/Tuliodna/status/1463911554538160130>.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

