Understanding Willow Transcriptional Response in the Context of Oil Sands Tailings Reclamation
- PMID: 35574135
- PMCID: PMC9094116
- DOI: 10.3389/fpls.2022.857535
Understanding Willow Transcriptional Response in the Context of Oil Sands Tailings Reclamation
Abstract
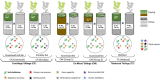
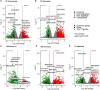
One of the reclamation objectives for treated oil sands tailings (OST) is to establish boreal forest communities that can integrate with the surrounding area. Hence, selection of appropriate soil reclamation cover designs and plant species for revegetation are important aspects of tailings landform reclamation and closure. Research and monitoring of the long term and immediate impacts of capped OST on the growth and survival of native boreal plant species are currently underway. However, plant responses to OST-associated toxicity are not well known at the molecular level. Using RNA sequencing, we examined the effects of three types of OST on the willow transcriptome under different capping strategies. The transcriptomic data showed that some genes respond universally and others in a specific manner to different types of OST. Among the dominant and shared upregulated genes, we found some encoding protein detoxification (PD), Cytochrome P450 (CYPs), glutathione S-transferase regulatory process (GST), UDP-glycosyltransferase (UGT), and ABC transporter and regulatory process associated proteins. Moreover, genes encoding several stress-responsive transcription factors (bZIP, BHLH, ERF, MYB, NAC, WRKY) were upregulated with OST-exposure, while high numbers of transcripts related to photosynthetic activity and chloroplast structure and function were downregulated. Overall, the expression of 40 genes was found consistent across all tailings types and capping strategies. The qPCR analysis of a subset of these shared genes suggested that they could reliably distinguish plants exposed to different OST associated stress. Our results indicated that it is possible to develop OST stress exposure biosensors merely based on changes in the level of expression of a relatively small set of genes. The outcomes of this study will further guide optimization of OST capping and revegetation technology by using knowledge based plant stress adaptation strategies.
Keywords: oil sands tailings; polycyclic aromatic hydrocarbon; reclamation; transcriptome; willow.
Copyright © 2022 Samad, Pelletier, Séguin, Degenhardt, Muench and Martineau.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Azam S., Rima U. S. (2014). Oil sand tailings characterisation for centrifuge dewatering. Environ. Geotech. 1 189–196. 10.1680/envgeo.13.00044 - DOI
-
- Badiozamani M. M., Askari-Nasab H. (2014). Integration of reclamation and tailings management in oil sands surface mine planning. Environ. Model. Softw. 51 45–58. 10.1016/j.envsoft.2013.09.026 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

