MET Inhibition Sensitizes Rhabdomyosarcoma Cells to NOTCH Signaling Suppression
- PMID: 35574376
- PMCID: PMC9092259
- DOI: 10.3389/fonc.2022.835642
MET Inhibition Sensitizes Rhabdomyosarcoma Cells to NOTCH Signaling Suppression
Abstract
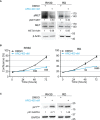
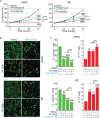
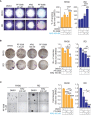
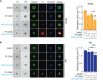
Rhabdomyosarcoma (RMS) is a pediatric myogenic soft tissue sarcoma. The Fusion-Positive (FP) subtype expresses the chimeric protein PAX3-FOXO1 (P3F) while the Fusion-Negative (FN) is devoid of any gene translocation. FP-RMS and metastatic FN-RMS are often unresponsive to conventional therapy. Therefore, novel therapeutic approaches are needed to halt tumor progression. NOTCH signaling has oncogenic functions in RMS and its pharmacologic inhibition through γ-secretase inhibitors blocks tumor growth in vitro and in vivo. Here, we show that NOTCH signaling blockade resulted in the up-regulation and phosphorylation of the MET oncogene in both RH30 (FP-RMS) and RD (FN-RMS) cell lines. Pharmacologic inhibition of either NOTCH or MET signaling slowed proliferation and restrained cell survival compared to control cells partly by increasing Annexin V and CASP3/7 activation. Co-treatment with NOTCH and MET inhibitors significantly amplified these effects and enhanced PARP1 cleavage in both cell lines. Moreover, it severely hampered cell migration, colony formation, and anchorage-independent growth compared to single-agent treatments in both cell lines and significantly prevented the growth of FN-RMS cells grown as spheroids. Collectively, our results unveil the overexpression of the MET oncogene by NOTCH signaling targeting in RMS cells and show that MET pathway blockade sensitizes them to NOTCH inhibition.
Keywords: MET; NOTCH signaling; combination therapy; drug resistance; rhabdomyosarcoma; soft tissue sarcoma; targeted therapy; γ-secretase.
Copyright © 2022 Perrone, Pomella, Cassandri, Pezzella, Milano, Colletti, Cossetti, Pericoli, Di Giannatale, de Billy, Vinci, Petrini, Marampon, Quintarelli, Taulli, Roma, Gallego, Camero, Mariottini, Cervelli, Maestro, Miele, De Angelis, Locatelli and Rota.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, et al. . Comprehensive Genomic Analysis of Rhabdomyosarcoma Reveals a Landscape of Alterations Affecting a Common Genetic Axis in Fusion-Positive and Fusion-Negative Tumors. Cancer Discov (2014) 4:216–31. doi: 10.1158/2159-8290.CD-13-0639 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

