Viral RNA Load and Infectivity of SARS-CoV-2 in Paired Respiratory and Oral Specimens from Symptomatic, Asymptomatic, or Postsymptomatic Individuals
- PMID: 35575498
- PMCID: PMC9241670
- DOI: 10.1128/spectrum.02264-21
Viral RNA Load and Infectivity of SARS-CoV-2 in Paired Respiratory and Oral Specimens from Symptomatic, Asymptomatic, or Postsymptomatic Individuals
Abstract
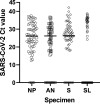
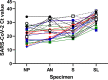
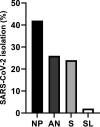
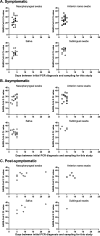
In the present study, we assessed the diagnostic sensitivity and determined the viral RNA load and infectivity of SARS-CoV-2 in paired respiratory (nasopharyngeal and anterior nares) and oral samples (saliva and sublingual swab). Samples were collected from 77 individuals of which 75 were diagnosed with COVID-19 and classified as symptomatic (n = 29), asymptomatic (n = 31), or postsymptomatic (n = 15). Specimens were collected at one time point from each individual, between day 1 and 23 after the initial COVID-19 diagnosis, and included self-collected saliva (S), or sublingual (SL) swab, and bilateral anterior nares (AN) swab, followed by health care provider collected nasopharyngeal (NP) swab. Sixty-three specimen sets were tested using five assay/platforms. The diagnostic sensitivity of each assay/platform and specimen type was determined. Of the 63 specimen sets, SARS-CoV-2 was detected in 62 NP specimens, 52 AN specimens, 59 saliva specimens, and 31 SL specimens by at least one platform. Infectious SARS-CoV-2 was isolated from 21 NP, 13 AN, 12 saliva, and one SL specimen out of 50 specimen sets. SARS-CoV-2 isolation was most successful up to 5 days after initial COVID-19 diagnosis using NP specimens from symptomatic patients (16 of 24 positives, 66.67%), followed by specimens from asymptomatic patients (5 of 17 positives, 29.41%), while it was not very successful with specimens from postsymptomatic patients. Benefits of self-collected saliva and AN specimens balance the loss of sensitivity relative to NP specimens. Therefore, saliva and AN specimens are acceptable alternatives for symptomatic SARS-CoV-2 diagnostic testing or surveillance with increased sampling frequency of asymptomatic individuals. IMPORTANCE The dynamics of infection with SARS-CoV-2 have a significant impact on virus infectivity and in the diagnostic sensitivity of molecular and classic virus detection tests. In the present study we determined the diagnostic sensitivity of paired respiratory (nasopharyngeal and anterior nares swabs) and oral secretions (saliva and sublingual swab) and assessed infectious virus shedding patterns by symptomatic, asymptomatic, or postsymptomatic individuals. Understanding the diagnostic performance of these specimens and the patterns of infectious virus shedding in these bodily secretions provides critical information to control COVID-19, and may help to refine guidelines on isolation and quarantine of positive individuals and their close contacts identified through epidemiological investigations.
Keywords: RT-PCR; SARS-CoV-2; anterior nares; diagnostic; saliva; virus isolation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. 2020. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med 382:1199–1207. doi: 10.1056/NEJMoa2001316. - DOI - PMC - PubMed
-
- Byrne AW, McEvoy D, Collins AB, Hunt K, Casey M, Barber A, Butler F, Griffin J, Lane EA, McAloon C, O'Brien K, Wall P, Walsh KA, More SJ. 2020. Inferred duration of infectious period of SARS-CoV-2: rapid scoping review and analysis of available evidence for asymptomatic and symptomatic COVID-19 cases. BMJ Open 10:e039856. doi: 10.1136/bmjopen-2020-039856. - DOI - PMC - PubMed
-
- He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, Lau YC, Wong JY, Guan Y, Tan X, Mo X, Chen Y, Liao B, Chen W, Hu F, Zhang Q, Zhong M, Wu Y, Zhao L, Zhang F, Cowling BJ, Li F, Leung GM. 2020. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 26:672–675. doi: 10.1038/s41591-020-0869-5. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

