Diversity and Long-Term Dynamics of Human Blood Anelloviruses
- PMID: 35575554
- PMCID: PMC9175625
- DOI: 10.1128/jvi.00109-22
Diversity and Long-Term Dynamics of Human Blood Anelloviruses
Abstract
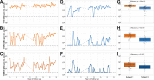
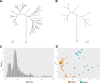
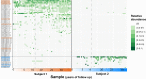
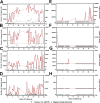
Anelloviruses (AVs) are commensal members of the human blood virome. Even though it was estimated that over 90% of the human population carries AVs, the dynamics of the AV virome ("anellome") are unknown. We investigated the dynamics of blood anellomes in two healthy people followed up for more than 30 years. Both subjects were positive for AVs in the majority of samples. Alphatorquevirus (torque teno virus [TTV]) was the most common genus in both subjects, followed by Betatorquevirus (torque teno minivirus [TTMV]) and Gammatorquevirus (torque teno midivirus [TTMDV]). Almost five times more lineages were found in subject 1 than in subject 2, and the anellomes differed phylogenetically. Both anellomes remained compositionally stable, and 9 out of 64 AV lineages were detected in over half of the time points. We confirmed the long-term and short-term persistence of 13 lineages by specific quantitative PCR (qPCR). AV lineages were detected in blood for over 30 years. Noticeable differences in anellome richness were found between the tested subjects, but both anellomes remained compositionally stable over time. These findings demonstrate that the human blood anellome is personal and that AV infection is chronic and potentially commensal. IMPORTANCE Knowledge of the persistence of AVs in humans is crucial to our understanding of the nature of AV infection (chronic or acute) and the role of AV in the host. We therefore investigated the dynamics of anellovirus infection in two healthy people followed up for 30 years. Our findings suggest that the human blood anellovirus virome (anellome) remains stable and personal for decades.
Keywords: anellome; anellovirus; blood virome; chronic viral infection; torque teno virus; virome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Vu D-L, Cordey S, Simonetta F, Brito F, Docquier M, Turin L, van Delden C, Boely E, Dantin C, Pradier A, Roosnek E, Chalandon Y, Zdobnov EM, Masouridi-Levrat S, Kaiser L. 2019. Human pegivirus persistence in human blood virome after allogeneic haematopoietic stem-cell transplantation. Clin Microbiol Infect 25:225–232. 10.1016/j.cmi.2018.05.004. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

