Spatiotemporal Heterogeneity and Intragenus Variability in Rhizobacterial Associations with Brassica rapa Growth
- PMID: 35575562
- PMCID: PMC9239066
- DOI: 10.1128/msystems.00060-22
Spatiotemporal Heterogeneity and Intragenus Variability in Rhizobacterial Associations with Brassica rapa Growth
Abstract
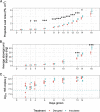
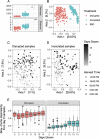
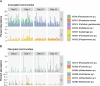
Microbial communities in the rhizosphere are distinct from those in soils and are influenced by stochastic and deterministic processes during plant development. These communities contain bacteria capable of promoting growth in host plants through various strategies. While some interactions are characterized in mechanistic detail using model systems, others can be inferred from culture-independent methods, such as 16S amplicon sequencing, using machine learning methods that account for this compositional data type. To characterize assembly processes and identify community members associated with plant growth amid the spatiotemporal variability of the rhizosphere, we grew Brassica rapa in a greenhouse time series with amended and reduced microbial treatments. Inoculation with a native soil community increased plant leaf area throughout the time series by up to 28%. Despite identifying spatially and temporally variable amplicon sequence variants (ASVs) in both treatments, inoculated communities were more highly connected and assembled more deterministically overall. Using a generalized linear modeling approach controlling for spatial variability, we identified 43 unique ASVs that were positively or negatively associated with leaf area, biomass, or growth rates across treatments and time stages. ASVs of the genus Flavobacterium dominated rhizosphere communities and showed some of the strongest positive and negative correlations with plant growth. Members of this genus, and growth-associated ASVs more broadly, exhibited variable connectivity in networks independent of growth association (positive or negative). These findings suggest host-rhizobacterial interactions vary temporally at narrow taxonomic scales and present a framework for identifying rhizobacteria that may work independently or in concert to improve agricultural yields. IMPORTANCE The rhizosphere, the zone of soil surrounding plant roots, is a hot spot for microbial activity, hosting bacteria capable of promoting plant growth in ways like increasing nutrient availability or fighting plant pathogens. This microbial system is highly diverse and most bacteria are unculturable, so to identify specific bacteria associated with plant growth, we used culture-independent community DNA sequencing combined with machine learning techniques. We identified 43 specific bacterial sequences associated with the growth of the plant Brassica rapa in different soil microbial treatments and at different stages of plant development. Most associations between bacterial abundances and plant growth were positive, although similar bacterial groups sometimes had different effects on growth. Why this happens will require more research, but overall, this study provides a way to identify native bacteria from plant roots that might be isolated and applied to boost agricultural yields.
Keywords: 16S RNA; Brassica rapa; feature selection; microbial communities; microbial community; plant growth promotion; rhizosphere; rhizosphere-inhabiting microbes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Marilley L, Aragno M. 1999. Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Appl Soil Ecol 13:127–136. doi:10.1016/S0929-1393(99)00028-1. - DOI
-
- Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P. 2012. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91–95. doi:10.1038/nature11336. - DOI - PubMed
-
- Harbort CJ, Hashimoto M, Inoue H, Niu Y, Guan R, Rombolà AD, Kopriva S, Voges M, Sattely ES, Garrido-Oter R, Schulze-Lefert P. 2020. Root-secreted coumarins and the microbiota interact to improve iron nutrition in Arabidopsis. Cell Host Microbe 28:825–837. doi:10.1016/j.chom.2020.09.006. - DOI - PMC - PubMed
-
- Rengel Z. 2015. Availability of Mn, Zn and Fe in the rhizosphere. J Soil Science Plant Nutrition 15:397–409. doi:10.4067/S0718-95162015005000036. - DOI
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
