Immune-Related Biomarkers Associated with Lung Metastasis from the Colorectal Cancer Microenvironment
- PMID: 35576491
- PMCID: PMC9142768
- DOI: 10.1089/jir.2021.0214
Immune-Related Biomarkers Associated with Lung Metastasis from the Colorectal Cancer Microenvironment
Abstract
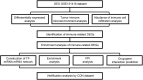
Immune-associated biomarkers can predict lung metastases from colorectal cancer. Differentially expressed genes (DEGs) were screened from sample data extracted from gene expression omnibus (GEO) database. The DEGs were screened from the lung metastasis (LM) and primary cancer (PC) groups of the Moffitt Cancer Center cohort dataset. Then, the tumor immune microenvironment and abundance of immune cell infiltration analyses were performed, and the immune-related DEGs were retrieved. In addition, the transcription factor (TF)-miRNA-mRNA network was constructed and enrichment analyses of the immune-related DEGs and upregulated and downregulated DEGs were carried out. Then, the protein-protein interaction (PPI) network was conducted and the drug-gene interaction was predicted. A total of 268 DEGs were screened. The Immune_Score of samples in the LM group was significantly higher compared with the PC group. The infiltration ratio of M0 macrophages and M2 macrophages of samples was higher than others. A total of 54 immune-related DEGs in M0 macrophages were screened. Moreover, the TF-miRNA-mRNA network was constructed among 8 miRNA-mRNA and 50 TF-mRNA, and the secreted phosphoprotein 1 was regulated by 12 TFs, and the oxidized low-density lipoprotein receptor 1 was regulated by 3 miRNAs and 3 TFs. The TF SAM pointed domain containing ETS TF was also a downregulated DEG. The Kyoto Encyclopedia of Genes and Genomes pathway analysis showed that the DEGs in the TF-miRNA-mRNA network were mainly involved in the interleukin-7 signaling pathway and cell adhesion molecules. In total, 23 protein interactions in this PPI network of M0 macrophage cells were involved in 27 mRNAs. There were 38 drug-gene interactions of immune-related DEGs of M0 macrophage cells predicted to contain 34 small molecule drugs and 8 mRNAs. Finally, the CON cohort dataset verified that the infiltration ratio of M0 and M2 macrophages of the samples was higher.
Keywords: colorectal cancer; differentially expressed genes; drug–gene interaction; immune cell infiltration; immune-related differentially expressed genes; lung metastasis.
Conflict of interest statement
No competing financial interests exist.
Figures






References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel TL, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene Ontology: tool for the unification of biology. Nucleic Acids Res 27(1):29–34. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

