Importance of Including Non-European Populations in Large Human Genetic Studies to Enhance Precision Medicine
- PMID: 35576557
- PMCID: PMC9904154
- DOI: 10.1146/annurev-biodatasci-122220-112550
Importance of Including Non-European Populations in Large Human Genetic Studies to Enhance Precision Medicine
Abstract
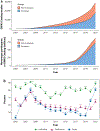
One goal of genomic medicine is to uncover an individual's genetic risk for disease, which generally requires data connecting genotype to phenotype, as done in genome-wide association studies (GWAS). While there may be clinical promise to employing prediction tools such as polygenic risk scores (PRS), it currently stands that individuals of non-European ancestry may not reap the benefits of genomic medicine because of underrepresentation in large-scale genetics studies. Here, we discuss why this inequity poses a problem for genomic medicine and the reasons for the low transferability of PRS across populations. We also survey the ancestry representation of published GWAS and investigate how estimates of ancestry diversity in GWASparticipants might be biased. We highlight the importance of expanding genetic research in Africa, one of the most underrepresented regions in human genomics research, and discuss issues of ethics, resources, and technology for equitable advancement of genomic medicine.
Keywords: Africa; GWAS; diversity; human genomics; polygenic risk score; precision medicine.
Figures


References
-
- Lawlor DA, Harbord RM, Sterne JAC, Timpson N, Davey Smith G. 2008. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Med 27(8):1133–63 - PubMed
-
- Sabatine MS. 2019. PCSK9 inhibitors: clinical evidence and implementation. Nat. Rev. Cardiol 16(3):155–65 - PubMed
-
- Frangoul H, Altshuler D, Cappellini MD, Chen Y-S, Domm J, et al. 2021. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N. Engl. J. Med 384(3):252–60 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

