OCA-T1 and OCA-T2 are coactivators of POU2F3 in the tuft cell lineage
- PMID: 35576971
- PMCID: PMC9419707
- DOI: 10.1038/s41586-022-04842-7
OCA-T1 and OCA-T2 are coactivators of POU2F3 in the tuft cell lineage
Abstract
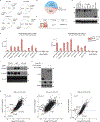
Tuft cells are a rare chemosensory lineage that coordinates immune and neural responses to foreign pathogens in mucosal tissues1. Recent studies have also revealed tuft-cell-like human tumours2,3, particularly as a variant of small-cell lung cancer. Both normal and neoplastic tuft cells share a genetic requirement for the transcription factor POU2F3 (refs. 2,4), although the transcriptional mechanisms that generate this cell type are poorly understood. Here we show that binding of POU2F3 to the uncharacterized proteins C11orf53 and COLCA2 (renamed here OCA-T1/POU2AF2 and OCA-T2/POU2AF3, respectively) is critical in the tuft cell lineage. OCA-T1 and OCA-T2 are paralogues of the B-cell-specific coactivator OCA-B; all three proteins are encoded in a gene cluster and contain a conserved peptide that binds to class II POU transcription factors and a DNA octamer motif in a bivalent manner. We demonstrate that binding between POU2F3 and OCA-T1 or OCA-T2 is essential in tuft-cell-like small-cell lung cancer. Moreover, we generated OCA-T1-deficient mice, which are viable but lack tuft cells in several mucosal tissues. These findings reveal that the POU2F3-OCA-T complex is the master regulator of tuft cell identity and a molecular vulnerability of tuft-cell-like small-cell lung cancer.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures












References
-
- Yamada Y et al. A tuft cell-like signature is highly prevalent in thymic squamous cell carcinoma and delineates new molecular subsets among the major lung cancer histotypes. J. Thorac. Oncol. 16, 1003–1016 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

