Characterization and antiviral susceptibility of SARS-CoV-2 Omicron BA.2
- PMID: 35576972
- PMCID: PMC10579982
- DOI: 10.1038/s41586-022-04856-1
Characterization and antiviral susceptibility of SARS-CoV-2 Omicron BA.2
Abstract
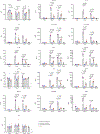
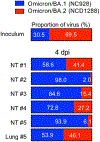
The recent emergence of SARS-CoV-2 Omicron (B.1.1.529 lineage) variants possessing numerous mutations has raised concerns of decreased effectiveness of current vaccines, therapeutic monoclonal antibodies and antiviral drugs for COVID-19 against these variants1,2. The original Omicron lineage, BA.1, prevailed in many countries, but more recently, BA.2 has become dominant in at least 68 countries3. Here we evaluated the replicative ability and pathogenicity of authentic infectious BA.2 isolates in immunocompetent and human ACE2-expressing mice and hamsters. In contrast to recent data with chimeric, recombinant SARS-CoV-2 strains expressing the spike proteins of BA.1 and BA.2 on an ancestral WK-521 backbone4, we observed similar infectivity and pathogenicity in mice and hamsters for BA.2 and BA.1, and less pathogenicity compared with early SARS-CoV-2 strains. We also observed a marked and significant reduction in the neutralizing activity of plasma from individuals who had recovered from COVID-19 and vaccine recipients against BA.2 compared to ancestral and Delta variant strains. In addition, we found that some therapeutic monoclonal antibodies (REGN10987 plus REGN10933, COV2-2196 plus COV2-2130, and S309) and antiviral drugs (molnupiravir, nirmatrelvir and S-217622) can restrict viral infection in the respiratory organs of BA.2-infected hamsters. These findings suggest that the replication and pathogenicity of BA.2 is similar to that of BA.1 in rodents and that several therapeutic monoclonal antibodies and antiviral compounds are effective against Omicron BA.2 variants.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures









Update of
-
Characterization and antiviral susceptibility of SARS-CoV-2 Omicron/BA.2.Res Sq [Preprint]. 2022 Feb 24:rs.3.rs-1375091. doi: 10.21203/rs.3.rs-1375091/v1. Res Sq. 2022. Update in: Nature. 2022 Jul;607(7917):119-127. doi: 10.1038/s41586-022-04856-1. PMID: 35233565 Free PMC article. Updated. Preprint.
References
-
- Callaway E & Ledford H How bad is Omicron? What scientists know so far. Nature 600, 197–199 (2021). - PubMed
-
- World Health Organization. Weekly epidemiological update on COVID-19, 5 April 2022, https://www.who.int/publications/m/item/weekly-epidemiological-update-on... (2022).
-
- Lyngse F et al. Transmission of SARS-CoV-2 Omicron VOC subvariants BA.1 and BA.2: evidence from Danish households. Preprint at medRxiv 10.1101/2022.01.28.22270044 (2022). - DOI
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

