Integrative structure determination reveals functional global flexibility for an ultra-multimodular arabinanase
- PMID: 35577850
- PMCID: PMC9110388
- DOI: 10.1038/s42003-022-03054-z
Integrative structure determination reveals functional global flexibility for an ultra-multimodular arabinanase
Abstract
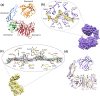
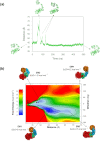
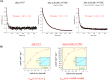
AbnA is an extracellular GH43 α-L-arabinanase from Geobacillus stearothermophilus, a key bacterial enzyme in the degradation and utilization of arabinan. We present herein its full-length crystal structure, revealing the only ultra-multimodular architecture and the largest structure to be reported so far within the GH43 family. Additionally, the structure of AbnA appears to contain two domains belonging to new uncharacterized carbohydrate-binding module (CBM) families. Three crystallographic conformational states are determined for AbnA, and this conformational flexibility is thoroughly investigated further using the "integrative structure determination" approach, integrating molecular dynamics, metadynamics, normal mode analysis, small angle X-ray scattering, dynamic light scattering, cross-linking, and kinetic experiments to reveal large functional conformational changes for AbnA, involving up to ~100 Å movement in the relative positions of its domains. The integrative structure determination approach demonstrated here may apply also to the conformational study of other ultra-multimodular proteins of diverse functions and structures.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Substrate cleavage pattern, biophysical characterization and low-resolution structure of a novel hyperthermostable arabinanase from Thermotoga petrophila.Biochem Biophys Res Commun. 2010 Sep 3;399(4):505-11. doi: 10.1016/j.bbrc.2010.07.097. Epub 2010 Aug 1. Biochem Biophys Res Commun. 2010. PMID: 20678476
-
Purification, characterization and functional analysis of an endo-arabinanase (AbnA) from Bacillus subtilis.FEMS Microbiol Lett. 2004 Dec 1;241(1):41-8. doi: 10.1016/j.femsle.2004.10.003. FEMS Microbiol Lett. 2004. PMID: 15556708
-
Probing the Complex Architecture of Multimodular Carbohydrate-Active Enzymes Using a Combination of Small Angle X-Ray Scattering and X-Ray Crystallography.Methods Mol Biol. 2017;1588:239-253. doi: 10.1007/978-1-4939-6899-2_19. Methods Mol Biol. 2017. PMID: 28417374
-
Structure and function analysis of Pseudomonas plant cell wall hydrolases.Prog Nucleic Acid Res Mol Biol. 1998;61:211-41. doi: 10.1016/s0079-6603(08)60828-4. Prog Nucleic Acid Res Mol Biol. 1998. PMID: 9752722 Review.
-
Structure and function of carbohydrate-binding module families 13 and 42 of glycoside hydrolases, comprising a β-trefoil fold.Biosci Biotechnol Biochem. 2013;77(7):1363-71. doi: 10.1271/bbb.130183. Epub 2013 Jul 7. Biosci Biotechnol Biochem. 2013. PMID: 23832347 Review.
Cited by
-
Interactions of the male contraceptive target EPPIN with semenogelin-1 and small organic ligands.Sci Rep. 2023 Sep 1;13(1):14382. doi: 10.1038/s41598-023-41365-1. Sci Rep. 2023. PMID: 37658081 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

