Inducing respiratory complex I impairment elicits an increase in PGC1α in ovarian cancer
- PMID: 35577908
- PMCID: PMC9110394
- DOI: 10.1038/s41598-022-11620-y
Inducing respiratory complex I impairment elicits an increase in PGC1α in ovarian cancer
Abstract
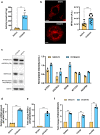
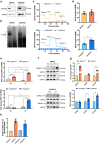
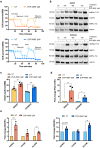
Anticancer strategies aimed at inhibiting Complex I of the mitochondrial respiratory chain are increasingly being attempted in solid tumors, as functional oxidative phosphorylation is vital for cancer cells. Using ovarian cancer as a model, we show that a compensatory response to an energy crisis induced by Complex I genetic ablation or pharmacological inhibition is an increase in the mitochondrial biogenesis master regulator PGC1α, a pleiotropic coactivator of transcription regulating diverse biological processes within the cell. We associate this compensatory response to the increase in PGC1α target gene expression, setting the basis for the comprehension of the molecular pathways triggered by Complex I inhibition that may need attention as drawbacks before these approaches are implemented in ovarian cancer care.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Brown JR, Chan DK, Shank JJ, Griffith KA, Fan H, Szulawski R, Yang K, Reynolds RK, Johnston C, McLean K, Uppal S, Liu JR, Cabrera L, Taylor SE, Orr BC, Modugno F, Mehta P, Bregenzer M, Mehta G, Shen H, Coffman L, Buckanovich RJ. Phase II clinical trial of metformin as a cancer stem cell-targeting agent in ovarian cancer. JCI Insight. 2020;5:e133247. doi: 10.1172/jci.insight.133247. - DOI - PMC - PubMed
-
- Molina JR, Sun Y, Protopopova M, Gera S, Bandi M, Bristow C, McAfoos T, Morlacchi P, Ackroyd J, Agip A-NA, Al-Atrash G, Asara J, Bardenhagen J, Carrillo CC, Carroll C, Chang E, Ciurea S, Cross JB, Czako B, Deem A, Daver N, de Groot JF, Dong J-W, Feng N, Gao G, Gay J, Do MG, Greer J, Giuliani V, Han J, Han L, Henry VK, Hirst J, Huang S, Jiang Y, Kang Z, Khor T, Konoplev S, Lin Y-H, Liu G, Lodi A, Lofton T, Ma H, Mahendra M, Matre P, Mullinax R, Peoples M, Petrocchi A, Rodriguez-Canale J, Serreli R, Shi T, Smith M, Tabe Y, Theroff J, Tiziani S, Xu Q, Zhang Q, Muller F, DePinho RA, Toniatti C, Draetta GF, Heffernan TP, Konopleva M, Jones P, Di Francesco ME, Marszalek JR. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nature Medicine. 2018;24(7):1036–1046. doi: 10.1038/s41591-018-0052-4. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

