Meta-matching as a simple framework to translate phenotypic predictive models from big to small data
- PMID: 35578132
- PMCID: PMC9202200
- DOI: 10.1038/s41593-022-01059-9
Meta-matching as a simple framework to translate phenotypic predictive models from big to small data
Abstract
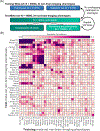
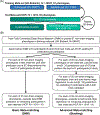
We propose a simple framework-meta-matching-to translate predictive models from large-scale datasets to new unseen non-brain-imaging phenotypes in small-scale studies. The key consideration is that a unique phenotype from a boutique study likely correlates with (but is not the same as) related phenotypes in some large-scale dataset. Meta-matching exploits these correlations to boost prediction in the boutique study. We apply meta-matching to predict non-brain-imaging phenotypes from resting-state functional connectivity. Using the UK Biobank (N = 36,848) and Human Connectome Project (HCP) (N = 1,019) datasets, we demonstrate that meta-matching can greatly boost the prediction of new phenotypes in small independent datasets in many scenarios. For example, translating a UK Biobank model to 100 HCP participants yields an eight-fold improvement in variance explained with an average absolute gain of 4.0% (minimum = -0.2%, maximum = 16.0%) across 35 phenotypes. With a growing number of large-scale datasets collecting increasingly diverse phenotypes, our results represent a lower bound on the potential of meta-matching.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
COMPETING INTERESTS STATEMENT
The authors declare no competing interests.
Figures






Comment in
-
Piggybacking on big data.Nat Neurosci. 2022 Jun;25(6):682-683. doi: 10.1038/s41593-022-01058-w. Nat Neurosci. 2022. PMID: 35578133 Free PMC article.
References
REFERENCES (MAIN TEXT)
-
- Varoquaux G & Poldrack RA Predictive models avoid excessive reductionism in cognitive neuroimaging. Curr. Opin. Neurobiol 55, 1–6 (2019). - PubMed
REFERENCES (METHODS)
-
- Varoquaux G et al. Assessing and tuning brain decoders: Cross-validation, caveats, and guidelines. Neuroimage 145, 166–179 (2017). - PubMed
-
- Tan C et al. A survey on deep transfer learning. Int. Conf. Artif. neural networks 11141 LNCS, 270–279 (2018).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

