Metagenomic Shotgun Sequencing of Endocervical, Vaginal, and Rectal Samples among Fijian Women with and without Chlamydia trachomatis Reveals Disparate Microbial Populations and Function across Anatomic Sites: a Pilot Study
- PMID: 35579443
- PMCID: PMC9241848
- DOI: 10.1128/spectrum.00105-22
Metagenomic Shotgun Sequencing of Endocervical, Vaginal, and Rectal Samples among Fijian Women with and without Chlamydia trachomatis Reveals Disparate Microbial Populations and Function across Anatomic Sites: a Pilot Study
Abstract
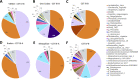
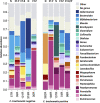
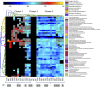
Chlamydia trachomatis is a sexually transmitted pathogen and a global public health concern. Little is known about the microbial composition and function across endocervical, vaginal, and rectal microbiomes in the context of C. trachomatis infection. We evaluated the microbiomes of 10 age-matched high-risk Fijian women with and without C. trachomatis using metagenomic shotgun sequencing (MSS). Lactobacillus iners and Lactobacillus crispatus dominated the vagina and endocervix of uninfected women. Species often found in higher relative abundance in bacterial vaginosis (BV)-Mageeibacillus indolicus, Prevotella spp., Sneathia spp., Gardnerella vaginalis, and Veillonellaceae spp.-were dominant in C. trachomatis-infected women. This combination of BV pathogens was unique to Pacific Islanders compared to previously studied groups. The C. trachomatis-infected endocervix had a higher diversity of microbiota and microbial profiles that were somewhat different from those of the vagina. However, community state type III (CST-III) and CST-IV predominated, reflecting pathogenic microbiota regardless of C. trachomatis infection status. Rectal microbiomes were dominated by Prevotella and Bacteroides, although four women had unique microbiomes with Gardnerella, Akkermansia, Bifidobacterium, and Brachyspira. A high level of microbial similarity across microbiomes in two C. trachomatis-infected women suggested intragenitorectal transmission. A number of metabolic pathways in the endocervix, driven by BV pathogens and C. trachomatis to meet nutritional requirements for survival/growth, 5-fold higher than that in the vagina indicated that endocervical microbial functions are likely more diverse and complex than those in the vagina. Our novel findings provide the impetus for larger prospective studies to interrogate microbial/microbiome interactions that promote C. trachomatis infection and better define the unique genitorectal microbiomes of Pacific Islanders. IMPORTANCE Chlamydia trachomatis is the primary cause of bacterial sexually transmitted infections worldwide, with a disturbing increase in annual rates. While there is a plethora of data on healthy and pathogenic vaginal microbiomes-defining microbial profiles and associations with sexually transmitted infections (STIs)-far fewer studies have similarly examined the endocervix or rectum. Further, vulnerable populations, such as Pacific Islanders, remain underrepresented in research. We investigated the microbial composition, structure, and function of these anatomic microbiomes using metagenomic shotgun sequencing among a Fijian cohort. We found, primarily among C. trachomatis-infected women, unique microbial profiles in endocervical, vaginal, and rectal microbiomes with an increased diversity and more complex microbial pathways in endocervical than vaginal microbiomes. Similarities in microbiome composition across sites for some women suggested intragenitorectal transmission. These novel insights into genitorectal microbiomes and their purported function require prospective studies to better define Pacific Islander microbiomes and microbial/microbiome interactions that promote C. trachomatis infection.
Keywords: Chlamydia trachomatis; endocervical microbiome; metabolomics; metagenomic shotgun sequencing; pathogenesis; rectal microbiome; sexually transmitted infections; vaginal microbiome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Unique microbial diversity, community composition, and networks among Pacific Islander endocervical and vaginal microbiomes with and without Chlamydia trachomatis infection in Fiji.mBio. 2024 Jan 16;15(1):e0306323. doi: 10.1128/mbio.03063-23. Epub 2023 Dec 20. mBio. 2024. PMID: 38117091 Free PMC article.
-
Molecular Identification of Cervical Microbes in HIV-Negative and HIV-Positive Women in an African Setting Using a Customized Bacterial Vaginosis Microbial DNA Quantitative PCR (qPCR) Array.Microbiol Spectr. 2022 Jun 29;10(3):e0222921. doi: 10.1128/spectrum.02229-21. Epub 2022 Jun 1. Microbiol Spectr. 2022. PMID: 35647888 Free PMC article.
-
Bacterial vaginosis-associated vaginal microbiota is an age-independent risk factor for Chlamydia trachomatis, Mycoplasma genitalium and Trichomonas vaginalis infections in low-risk women, St. Petersburg, Russia.Eur J Clin Microbiol Infect Dis. 2020 Jul;39(7):1221-1230. doi: 10.1007/s10096-020-03831-w. Epub 2020 Feb 8. Eur J Clin Microbiol Infect Dis. 2020. PMID: 32036466 Free PMC article.
-
The two-sided role of the vaginal microbiome in Chlamydia trachomatis and Mycoplasma genitalium pathogenesis.J Reprod Immunol. 2018 Nov;130:11-17. doi: 10.1016/j.jri.2018.08.006. Epub 2018 Aug 22. J Reprod Immunol. 2018. PMID: 30149363 Review.
-
[Characteristics and physiologic role of female lower genital microbiome].Orv Hetil. 2023 Jun 18;164(24):923-930. doi: 10.1556/650.2023.32791. Print 2023 Jun 18. Orv Hetil. 2023. PMID: 37330978 Review. Hungarian.
Cited by
-
All Properties of Infertility Microbiome in a Review Article.J Clin Lab Anal. 2025 Mar;39(6):e25158. doi: 10.1002/jcla.25158. Epub 2025 Mar 9. J Clin Lab Anal. 2025. PMID: 40059472 Free PMC article. Review.
-
In-Silico Functional Metabolic Pathways Associated to Chlamydia trachomatis Genital Infection.Int J Mol Sci. 2022 Dec 13;23(24):15847. doi: 10.3390/ijms232415847. Int J Mol Sci. 2022. PMID: 36555488 Free PMC article.
-
Lactobacillus gasseri LGV03-derived indole-3-lactic acid ameliorates immune response by activating aryl hydrocarbon receptor.Microb Cell Fact. 2025 Jan 30;24(1):34. doi: 10.1186/s12934-025-02662-8. Microb Cell Fact. 2025. PMID: 39885499 Free PMC article.
-
A manifold-based framework for studying the dynamics of the vaginal microbiome.NPJ Biofilms Microbiomes. 2023 Dec 15;9(1):102. doi: 10.1038/s41522-023-00471-8. NPJ Biofilms Microbiomes. 2023. PMID: 38102172 Free PMC article.
-
Crosstalk between the Resident Microbiota and the Immune Cells Regulates Female Genital Tract Health.Life (Basel). 2023 Jul 9;13(7):1531. doi: 10.3390/life13071531. Life (Basel). 2023. PMID: 37511906 Free PMC article. Review.
References
-
- WHO. 2018. Report on global sexually transmitted infection surveillance. World Health Organization, Geneva, Switzerland.
-
- CDC. 2019. Sexually transmitted disease surveillance. Centers for Disease Control and Prevention, Atlanta, GA.
-
- WHO Regional Office for the Western Pacific. 2012. WHO multi-country cooperation strategy for the Pacific 2013–2017. WHO Regional Office for the Western Pacific, Manila, Philippines.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

