The Novel Amidase PcnH Initiates the Degradation of Phenazine-1-Carboxamide in Sphingomonas histidinilytica DS-9
- PMID: 35579476
- PMCID: PMC9195955
- DOI: 10.1128/aem.00543-22
The Novel Amidase PcnH Initiates the Degradation of Phenazine-1-Carboxamide in Sphingomonas histidinilytica DS-9
Abstract
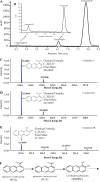
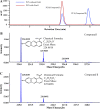
Phenazines are an important class of secondary metabolites and are primarily named for their heterocyclic phenazine cores, including phenazine-1-carboxylic acid (PCA) and its derivatives, such as phenazine-1-carboxamide (PCN) and pyocyanin (PYO). Although several genes involved in the degradation of PCA and PYO have been reported so far, the genetic foundations of PCN degradation remain unknown. In this study, a PCN-degrading bacterial strain, Sphingomonas histidinilytica DS-9, was isolated. The gene pcnH, encoding a novel amidase responsible for the initial step of PCN degradation, was cloned by genome comparison and subsequent experimental validation. PcnH catalyzed the hydrolysis of the amide bond of PCN to produce PCA, which shared low identity (only 26 to 33%) with reported amidases. The Km and kcat values of PcnH for PCN were 33.22 ± 5.70 μM and 18.71 ± 0.52 s-1, respectively. PcnH has an Asp-Lys-Cys motif, which is conserved among amidases of the isochorismate hydrolase-like (IHL) superfamily. The replacement of Asp37, Lys128, and Cys163 with alanine in PcnH led to the complete loss of enzymatic activity. Furthermore, the genes pcaA1A2A3A4 and pcnD were found to encode PCA 1,2-dioxygenase and 1,2-dihydroxyphenazine (2OHPC) dioxygenase, which were responsible for the subsequent degradation steps of PCN. The PCN-degradative genes were highly conserved in some bacteria of the genus Sphingomonas, with slight variations in the sequence identities. IMPORTANCE Phenazines have been widely acknowledged as a natural antibiotic for more than 150 years, but their degradation mechanisms are still not completely elucidated. Compared with the studies on the degradation mechanism of PCA and PYO, little is known regarding PCN degradation by far. Previous studies have speculated that its initial degradation step may be catalyzed by an amidase, but no further studies have been conducted. This study identified a novel amidase, PcnH, that catalyzed the hydrolysis of PCN to PCA. In addition, the PCA 1,2-dioxygenase PcaA1A2A3A4 and 2OHPC dioxygenase PcnD were also found to be involved in the subsequent degradation steps of PCN in S. histidinilytica DS-9. And the genes responsible for PCN catabolism are highly conserved in some strains of Sphingomonas. These results deepen our understanding of the PCN degradation mechanism.
Keywords: Sphingomonas histidinilytica DS-9; amidase PcnH; degradation; phenazine-1-carboxamide.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

