A translatable RNAi-driven gene therapy silences PMP22/Pmp22 genes and improves neuropathy in CMT1A mice
- PMID: 35579942
- PMCID: PMC9246392
- DOI: 10.1172/JCI159814
A translatable RNAi-driven gene therapy silences PMP22/Pmp22 genes and improves neuropathy in CMT1A mice
Abstract
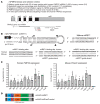
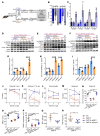
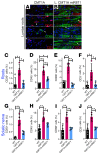
Charcot-Marie-Tooth disease type 1A (CMT1A), the most common inherited demyelinating peripheral neuropathy, is caused by PMP22 gene duplication. Overexpression of WT PMP22 in Schwann cells destabilizes the myelin sheath, leading to demyelination and ultimately to secondary axonal loss and disability. No treatments currently exist that modify the disease course. The most direct route to CMT1A therapy will involve reducing PMP22 to normal levels. To accomplish this, we developed a gene therapy strategy to reduce PMP22 using artificial miRNAs targeting human PMP22 and mouse Pmp22 mRNAs. Our lead therapeutic miRNA, miR871, was packaged into an adeno-associated virus 9 (AAV9) vector and delivered by lumbar intrathecal injection into C61-het mice, a model of CMT1A. AAV9-miR871 efficiently transduced Schwann cells in C61-het peripheral nerves and reduced human and mouse PMP22 mRNA and protein levels. Treatment at early and late stages of the disease significantly improved multiple functional outcome measures and nerve conduction velocities. Furthermore, myelin pathology in lumbar roots and femoral motor nerves was ameliorated. The treated mice also showed reductions in circulating biomarkers of CMT1A. Taken together, our data demonstrate that AAV9-miR871-driven silencing of PMP22 rescues a CMT1A model and provides proof of principle for treating CMT1A using a translatable gene therapy approach.
Keywords: Gene therapy; Mouse models; Neurodegeneration; Neuroscience; Therapeutics.
Conflict of interest statement
Figures





References
-
- Skre H. Genetic and clinical aspects of Charcot-Marie-Tooth’s disease. Clin Genet. 1974;6(2):98–118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

