Cancer driver drug interaction explorer
- PMID: 35580047
- PMCID: PMC9252786
- DOI: 10.1093/nar/gkac384
Cancer driver drug interaction explorer
Abstract
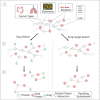
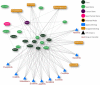
Cancer is a heterogeneous disease characterized by unregulated cell growth and promoted by mutations in cancer driver genes some of which encode suitable drug targets. Since the distinct set of cancer driver genes can vary between and within cancer types, evidence-based selection of drugs is crucial for targeted therapy following the precision medicine paradigm. However, many putative cancer driver genes can not be targeted directly, suggesting an indirect approach that considers alternative functionally related targets in the gene interaction network. Once potential drug targets have been identified, it is essential to consider all available drugs. Since tools that offer support for systematic discovery of drug repurposing candidates in oncology are lacking, we developed CADDIE, a web application integrating six human gene-gene and four drug-gene interaction databases, information regarding cancer driver genes, cancer-type specific mutation frequencies, gene expression information, genetically related diseases, and anticancer drugs. CADDIE offers access to various network algorithms for identifying drug targets and drug repurposing candidates. It guides users from the selection of seed genes to the identification of therapeutic targets or drug candidates, making network medicine algorithms accessible for clinical research. CADDIE is available at https://exbio.wzw.tum.de/caddie/ and programmatically via a python package at https://pypi.org/project/caddiepy/.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Nogales C., Mamdouh Z.M., List M., Kiel C., Casas A.I., Schmidt H.H.. Network pharmacology: curing causal mechanisms instead of treating symptoms. Trends Pharmacol. Sci. 2022; 43:136–150. - PubMed
-
- Rubin E.H., Gilliland D.G.. Drug development and clinical trials—the path to an approved cancer drug. Nat. Rev. Clin. Oncol. 2012; 9:215–222. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

