Soluble HLA peptidome of pleural effusions is a valuable source for tumor antigens
- PMID: 35580925
- PMCID: PMC9114951
- DOI: 10.1136/jitc-2021-003733
Soluble HLA peptidome of pleural effusions is a valuable source for tumor antigens
Abstract
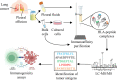
Background: Soluble human leucocyte antigen (sHLA) molecules, released into the plasma, carry their original peptide cargo and provide insight into the protein synthesis and degradation schemes of their source cells and tissues. Other body fluids, such as pleural effusions, may also contain sHLA-peptide complexes, and can potentially serve as a source of tumor antigens since these fluids are drained from the tumor microenvironment. We explored this possibility by developing a methodology for purifying and analyzing large pleural effusion sHLA class I peptidomes of patients with malignancies or benign diseases.
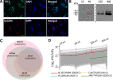
Methods: Cleared pleural fluids, cell pellets present in the pleural effusions, and the primary tumor cells cultured from cancer patients' effusions, were used for immunoaffinity purification of the HLA molecules. The recovered HLA peptides were analyzed by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) and the resulting LC-MS/MS data were analyzed with the MaxQuant software tool. Selected tumor antigen peptides were tested for their immunogenicity potential with donor peripheral blood mononuclear cells (PBMCs) in an in vitro assay.
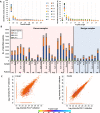
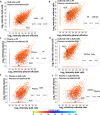
Results: Mass spectrometry analysis of the pleural effusions revealed 39,669 peptides attributable to 11,305 source proteins. The majority of peptides identified from the pleural effusions were defined as HLA ligands that fit the patients' HLA consensus sequence motifs. The membranal and soluble HLA peptidomes of each individual patient correlated to each other. Additionally, soluble HLA peptidomes from the same patient, obtained at different visits to the clinic, were highly similar. Compared with benign effusions, the soluble HLA peptidomes of malignant pleural effusions were larger and included HLA peptides derived from known tumor-associated antigens, including cancer/testis antigens, lung-related proteins, and vascular endothelial growth factor pathway proteins. Selected tumor-associated antigens that were identified by the immunopeptidomics were able to successfully prime CD8+ T cells.
Conclusions: Pleural effusions contain sHLA-peptide complexes, and the pleural effusion HLA peptidome of patients with malignant tumors can serve as a rich source of biomarkers for tumor diagnosis and potential candidates for personalized immunotherapy.
Keywords: antigen presentation; antigens; immunotherapy; lung neoplasms; tumor biomarkers.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures

Similar articles
-
Identification of Tumor Antigens Among the HLA Peptidomes of Glioblastoma Tumors and Plasma.Mol Cell Proteomics. 2019 Jun;18(6):1255-1268. doi: 10.1074/mcp.RA119.001524. Mol Cell Proteomics. 2019. PMID: 31154438 Free PMC article.
-
Soluble plasma HLA peptidome as a potential source for cancer biomarkers.Proc Natl Acad Sci U S A. 2010 Nov 2;107(44):18769-76. doi: 10.1073/pnas.1008501107. Epub 2010 Oct 25. Proc Natl Acad Sci U S A. 2010. PMID: 20974924 Free PMC article.
-
Identification of Tumor Antigens Among the HLA Peptidomes of Glioblastoma Tumors and Plasma.Mol Cell Proteomics. 2018 Nov;17(11):2132-2145. doi: 10.1074/mcp.RA118.000792. Epub 2018 Aug 2. Mol Cell Proteomics. 2018. Retracted and republished in: Mol Cell Proteomics. 2019 Jun;18(6):1255-1268. doi: 10.1074/mcp.RA119.001524. Retraction in: Mol Cell Proteomics. 2019 Jun;18(6):1270. doi: 10.1074/mcp.W119.001571. PMID: 30072578 Free PMC article. Retracted and republished. Retracted.
-
Soluble HLA class I molecules/CD8 ligation trigger apoptosis of CD8+ cells by Fas/Fas-ligand interaction.ScientificWorldJournal. 2002 Feb 12;2:421-3. doi: 10.1100/tsw.2002.122. ScientificWorldJournal. 2002. PMID: 12806026 Free PMC article. Review.
-
The Potential of Soluble Human Leukocyte Antigen Molecules for Early Cancer Detection and Therapeutic Vaccine Design.Vaccines (Basel). 2020 Dec 18;8(4):775. doi: 10.3390/vaccines8040775. Vaccines (Basel). 2020. PMID: 33353014 Free PMC article. Review.
Cited by
-
Multiple Classes of Antigen Contribute to the Antigenic Landscape of Mesothelioma.Mol Cell Proteomics. 2025 Mar;24(3):100925. doi: 10.1016/j.mcpro.2025.100925. Epub 2025 Feb 5. Mol Cell Proteomics. 2025. PMID: 39921204 Free PMC article.
-
Soluble HLA peptidome: A new resource for cancer biomarkers.Front Oncol. 2022 Dec 22;12:1069635. doi: 10.3389/fonc.2022.1069635. eCollection 2022. Front Oncol. 2022. PMID: 36620582 Free PMC article. Review.
-
SAPrIm 2.0: a semi-automated protocol for mid-throughput soluble HLA immunopeptidomics.Front Immunol. 2025 Apr 24;16:1546629. doi: 10.3389/fimmu.2025.1546629. eCollection 2025. Front Immunol. 2025. PMID: 40342411 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
