Hepatic p63 regulates glucose metabolism by repressing SIRT1
- PMID: 35580962
- PMCID: PMC9933162
- DOI: 10.1136/gutjnl-2021-326620
Hepatic p63 regulates glucose metabolism by repressing SIRT1
Abstract
Objective: p63 is a transcription factor within the p53 protein family that has key roles in development, differentiation and prevention of senescence, but its metabolic actions remain largely unknown. Herein, we investigated the physiological role of p63 in glucose metabolism.
Design: We used cell lines and mouse models to genetically manipulate p63 in hepatocytes. We also measured p63 in the liver of patients with obesity with or without type 2 diabetes (T2D).
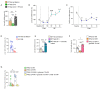
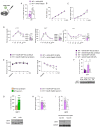
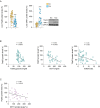
Results: We show that hepatic p63 expression is reduced on fasting. Mice lacking the specific isoform TAp63 in the liver (p63LKO) display higher postprandial and pyruvate-induced glucose excursions. These mice have elevated SIRT1 levels, while SIRT1 knockdown in p63LKO mice normalises glycaemia. Overexpression of TAp63 in wild-type mice reduces postprandial, pyruvate-induced blood glucose and SIRT1 levels. Studies carried out in hepatocyte cell lines show that TAp63 regulates SIRT1 promoter by repressing its transcriptional activation. TAp63 also mediates the inhibitory effect of insulin on hepatic glucose production, as silencing TAp63 impairs insulin sensitivity. Finally, protein levels of TAp63 are reduced in obese persons with T2D and are negatively correlated with fasting glucose and homeostasis model assessment index.
Conclusions: These results demonstrate that p63 physiologically regulates glucose homeostasis.
Keywords: diabetes mellitus; diet; glucose metabolism; liver.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures




Comment in
-
Transcription factor p63, a member of the p53 family of tumour suppressors, regulates hepatic glucose metabolism.Gut. 2023 Mar;72(3):415-416. doi: 10.1136/gutjnl-2022-327790. Epub 2022 Jun 15. Gut. 2023. PMID: 35705366 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
