Helical Indexing in Real Space
- PMID: 35581231
- PMCID: PMC9114412
- DOI: 10.1038/s41598-022-11382-7
Helical Indexing in Real Space
Abstract
Biological structures with helical symmetries of distinct twist, rise, and axial symmetry are abundant and span a wide range of organisms and functions. Performing de novo helical indexing remains challenging because of the steep learning curve involved in Fourier space layer lines. The unknown amount of out-of-plane tilt and the existence of multiple conformations of the helices further complicate indexing. In this work, we introduce a real-space indexing method that leverages the prior knowledge of the tilt and in-plane angles of the helical filaments/tubes, robust ab initio 3D reconstruction capabilities in single particle cryo-EM to obtain asymmetric reconstructions, and automatic indexing of helical parameters directly from the asymmetric density maps. We validated this approach using data from multiple helical structures of distinct helical symmetries, diameters, flexibility, data qualities, and heterogeneous states. The fully automated tool we introduce for real space indexing, HI3D, uses the 2D lattice in the autocorrelation of the cylindrical projection of a 3D density map to identify the helical symmetry. HI3D can often successfully determine the helical parameters of a suboptimal 3D density map, including ab initio single particle asymmetric reconstructions and sub-tomogram averages, with intermediate evidence that can also help assess the map quality. Furthermore, this open-source HI3D is usable independently as a Web application that can be accessed free of installation. With these methods, de novo helical indexing will be significantly more accessible to researchers investigating structures of helical filaments/tubes using cryo-EM.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

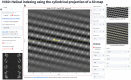
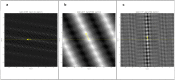






References
-
- Kidd, M. Paired helical filaments in electron microscopy of alzheimer’s disease (1963). - PubMed
-
- Terry, R. D. The fine structure of neurofibrillary tangles in Alzheimer’s disease (1963). - PubMed
-
- De Rosier, D. J. & Klug, A. Reconstruction of three dimensional structures from electron micrographs (1968). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

