An immune gene signature to predict prognosis and immunotherapeutic response in lung adenocarcinoma
- PMID: 35581376
- PMCID: PMC9114138
- DOI: 10.1038/s41598-022-12301-6
An immune gene signature to predict prognosis and immunotherapeutic response in lung adenocarcinoma
Abstract
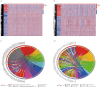
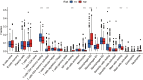
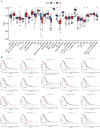
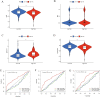
Lung adenocarcinoma is one of the most common malignant tumors worldwide. The purpose of this study was to construct a stable immune gene signature for prediction of prognosis (IGSPP) and response to immune checkpoint inhibitors (ICIs) therapy in LUAD patients. Five genes were screened by weighted gene coexpression network analysis, Cox regression and LASSO regression analyses and were used to construct the IGSPP. The survival rate of the IGSPP low-risk group was higher than that of the IGSPP high-risk group. Multivariate Cox regression analysis showed that IGSPP could be used as an independent prognostic factor for the overall survival of LUAD patients. IGSPP genes were enriched in cell cycle pathways. IGSPP gene mutation rates were higher in the high-risk group. CD4 memory-activated T cells, M0 and M1 macrophages had higher infiltration abundance in the high-risk group, which was associated with poor overall survival. In contrast, the abundance of resting CD4 memory T cells, monocytes, resting dendritic cells and resting mast cells associated with a better prognosis was higher in the low-risk group. TIDE scores and the expressions of different immune checkpoints showed that patients in the high-risk IGSPP group benefited more from ICIs treatment. In short, an IGSPP of LUAD was constructed and characterized. It could be used to predict the prognosis and benefits of ICIs treatment in LUAD patients.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Reck M, et al. Atezolizumab in combination with bevacizumab, paclitaxel and carboplatin for the first-line treatment of patients with metastatic non-squamous non-small cell lung cancer, including patients with EGFR mutations. Expert Rev. Respir. Med. 2020;14:125–136. doi: 10.1080/17476348.2020.1701439. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

