RHAMNETIN IS A BETTER INHIBITOR OF SARS-COV-2 2'-O-METHYLTRANSFERASE THAN DOLUTEGRAVIR: A COMPUTATIONAL PREDICTION
- PMID: 35582066
- PMCID: PMC9097307
- DOI: 10.21010/Ajid.v16i2.9
RHAMNETIN IS A BETTER INHIBITOR OF SARS-COV-2 2'-O-METHYLTRANSFERASE THAN DOLUTEGRAVIR: A COMPUTATIONAL PREDICTION
Abstract
Background: The 2'-O-methyltransferase is responsible for the capping of SARS-CoV-2 mRNA and consequently the evasion of the host's immune system. This study aims at identifying prospective natural inhibitors of the active site of SARS-CoV-2 2'O-methyltransferase (2'-OMT) through an in silico approach.
Materials and methods: The target was docked against a library of natural compounds obtained from edible African plants using PyRx - virtual screening software. The antiviral agent, Dolutegravir which has a binding affinity score of -8.5 kcal mol-1 with the SARS-CoV-2 2'-OMT was used as a standard. Compounds were screened for bioavailability through the SWISSADME web server using their molecular descriptors. Screenings for pharmacokinetic properties and bioactivity were performed with PKCSM and Molinspiration web servers respectively. The PLIP and Fpocket webservers were used for the binding site analyses. The Galaxy webserver was used for simulating the time-resolved motions of the apo and holo forms of the target while the MDWeb web server was used for the analyses of the trajectory data.
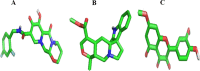
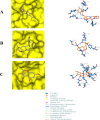
Results: The Root-Mean-Square-Deviation (RMSD) induced by Rhamnetin is 1.656A0 compared to Dolutegravir (1.579A0). The average B-factor induced by Rhamnetin is 113.75 while for Dolutegravir is 78.87; the Root-Mean-Square-Fluctuation (RMSF) for Rhamnetin is 0.75 and for Dolutegravir is 0.67. Also, at the active site, Rhamnetin also has a binding affinity score of -9.5 kcal mol-1 and forms 7 hydrogen bonds compared to Dolutegravir which has -8.5 kcal mol-1 and forms 4 hydrogen bonds respectively.
Conclusion: Rhamnetin showed better inhibitory activity at the target's active site than Dolutegravir.
Keywords: 2’-O-methyltransferase Inhibition; COVID-19; Computational Drug prediction; Coronavirus disease; SARS-CoV-2.
Copyright: © 2022 Afr. J. Infect. Diseases.
Figures
Similar articles
-
Identifying potential natural inhibitors of Brucella melitensis Methionyl-tRNA synthetase through an in-silico approach.PLoS Negl Trop Dis. 2022 Mar 21;16(3):e0009799. doi: 10.1371/journal.pntd.0009799. eCollection 2022 Mar. PLoS Negl Trop Dis. 2022. PMID: 35312681 Free PMC article.
-
Camptothecin shows better promise than Curcumin in the inhibition of the Human Telomerase: A computational study.Heliyon. 2021 Aug 10;7(8):e07742. doi: 10.1016/j.heliyon.2021.e07742. eCollection 2021 Aug. Heliyon. 2021. PMID: 34485722 Free PMC article.
-
In silico validation of coumarin derivatives as potential inhibitors against Main Protease, NSP10/NSP16-Methyltransferase, Phosphatase and Endoribonuclease of SARS CoV-2.J Biomol Struct Dyn. 2021 Nov;39(18):7306-7321. doi: 10.1080/07391102.2020.1808075. Epub 2020 Aug 24. J Biomol Struct Dyn. 2021. PMID: 32835632 Free PMC article.
-
Structure-based virtual screening and molecular dynamics simulation of SARS-CoV-2 Guanine-N7 methyltransferase (nsp14) for identifying antiviral inhibitors against COVID-19.J Biomol Struct Dyn. 2021 Aug;39(13):4582-4593. doi: 10.1080/07391102.2020.1778535. Epub 2020 Jun 22. J Biomol Struct Dyn. 2021. PMID: 32567979 Free PMC article.
-
Identification of a novel inhibitor of SARS-CoV-2 3CL-PRO through virtual screening and molecular dynamics simulation.PeerJ. 2021 Apr 13;9:e11261. doi: 10.7717/peerj.11261. eCollection 2021. PeerJ. 2021. PMID: 33954055 Free PMC article.
Cited by
-
Identification and characterization of alternative sites and molecular probes for SARS-CoV-2 target proteins.Front Chem. 2022 Oct 31;10:1017394. doi: 10.3389/fchem.2022.1017394. eCollection 2022. Front Chem. 2022. PMID: 36385993 Free PMC article.
-
Identifying immunodominant multi-epitopes from the envelope glycoprotein of the Lassa mammarenavirus as vaccine candidate for Lassa fever.Clin Exp Vaccine Res. 2022 Sep;11(3):249-263. doi: 10.7774/cevr.2022.11.3.249. Epub 2022 Sep 30. Clin Exp Vaccine Res. 2022. PMID: 36451670 Free PMC article.
References
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS:High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1. 2015;1:19–25.
-
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Grüning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses. Nucleic Acids Research, 2. 2018;46(W1):W537–W544. doi:10.1093/nar/gky379. PMID:29790989;PMCID:PMC6030816. - PMC - PubMed
-
- Al Wasidi AS, Hassan AS, Naglah AM. In vitro cytotoxicity and druglikeness of pyrazolines and pyridines bearing benzofuran moiety. Journal of Applied Pharmaceutical Science. 2020;10(04):142–148.
LinkOut - more resources
Full Text Sources
Miscellaneous
