Analyzing single cell transcriptome data from severe COVID-19 patients
- PMID: 35582459
- PMCID: PMC9021126
- DOI: 10.1016/j.xpro.2022.101379
Analyzing single cell transcriptome data from severe COVID-19 patients
Abstract
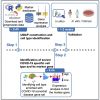
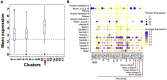
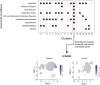
We describe the protocol for identifying COVID-19 severity specific cell types and their regulatory marker genes using single-cell transcriptomics data. We construct COVID-19 comorbid disease-associated gene list using multiple databases and literature resources. Next, we identify specific cell type where comorbid genes are upregulated. We further characterize the identified cell type using gene enrichment analysis. We detect upregulation of marker gene restricted to severe COVID-19 cell type and validate our findings using in silico, in vivo, and in vitro cellular models. For complete details on the use and execution of this protocol, please refer to Nassir et al. (2021b).
Keywords: Bioinformatics; Gene Expression; Genomics; Health Sciences; Immunology; Molecular Biology; RNAseq.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O., et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. - DOI - PMC - PubMed
-
- Bhattacharya S., Dunn P., Thomas C.G., Smith B., Schaefer H., Chen J., Hu Z., Zalocusky K.A., Shankar R.D., Shen-Orr S.S., et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci. Data. 2018;5:180015. doi: 10.1038/sdata.2018.15. - DOI - PMC - PubMed
-
- Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

