OsJAZ11 regulates spikelet and seed development in rice
- PMID: 35582630
- PMCID: PMC9090556
- DOI: 10.1002/pld3.401
OsJAZ11 regulates spikelet and seed development in rice
Abstract
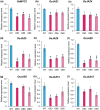
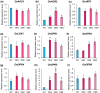
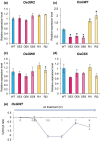
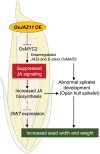
Seed size is one of the major determinants of seed weight and eventually, crop yield. As the global population is increasing beyond the capacity of current food production, enhancing seed size is a key target for crop breeders. Despite the identification of several genes and QTLs, current understanding about the molecular regulation of seed size/weight remains fragmentary. In the present study, we report novel role of a jasmonic acid (JA) signaling repressor, OsJAZ11 controlling rice seed width and weight. Transgenic rice lines overexpressing OsJAZ11 exhibited up to a 14% increase in seed width and ~30% increase in seed weight compared to wild type (WT). Constitutive expression of OsJAZ11 dramatically influenced spikelet morphogenesis leading to extra glume-like structures, open hull, and abnormal numbers of floral organs. Furthermore, overexpression lines accumulated higher JA levels in spikelets and developing seeds. Expression studies uncovered altered expression of JA biosynthesis/signaling and MADS box genes in overexpression lines compared to WT. Yeast two-hybrid and pull-down assays revealed that OsJAZ11 interacts with OsMADS29 and OsMADS68. Remarkably, expression of OsGW7, a key negative regulator of grain size, was significantly reduced in overexpression lines. We propose that OsJAZ11 participates in the regulation of seed size and spikelet development by coordinating the expression of JA-related, OsGW7 and MADS genes.
Keywords: JASMONATE ZIM‐DOMAIN; Jasmonic acid; seed weight; seed width.
© 2022 The Authors. Plant Direct published by American Society of Plant Biologists and the Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
OsJAZ11 regulates phosphate starvation responses in rice.Planta. 2021 Jun 18;254(1):8. doi: 10.1007/s00425-021-03657-6. Planta. 2021. PMID: 34143292 Free PMC article.
-
Jasmonic acid regulates spikelet development in rice.Nat Commun. 2014 Mar 19;5:3476. doi: 10.1038/ncomms4476. Nat Commun. 2014. PMID: 24647160
-
Cyanobacteria Application Ameliorates Floral Traits and Outcrossing Rate in Diverse Rice Cytoplasmic Male Sterile Lines.Plants (Basel). 2022 Dec 7;11(24):3411. doi: 10.3390/plants11243411. Plants (Basel). 2022. PMID: 36559523 Free PMC article.
-
Genes and Their Molecular Functions Determining Seed Structure, Components, and Quality of Rice.Rice (N Y). 2022 Mar 18;15(1):18. doi: 10.1186/s12284-022-00562-8. Rice (N Y). 2022. PMID: 35303197 Free PMC article. Review.
-
Effect of Panicle Morphology on Grain Filling and Rice Yield: Genetic Control and Molecular Regulation.Front Genet. 2022 May 10;13:876198. doi: 10.3389/fgene.2022.876198. eCollection 2022. Front Genet. 2022. PMID: 35620460 Free PMC article. Review.
Cited by
-
Developing Genetic Engineering Techniques for Control of Seed Size and Yield.Int J Mol Sci. 2022 Oct 31;23(21):13256. doi: 10.3390/ijms232113256. Int J Mol Sci. 2022. PMID: 36362043 Free PMC article. Review.
-
Natural allelic variation in GRAIN SIZE AND WEIGHT 3 of wild rice regulates the grain size and weight.Plant Physiol. 2023 Aug 31;193(1):502-518. doi: 10.1093/plphys/kiad320. Plant Physiol. 2023. PMID: 37249047 Free PMC article.
-
Identification and Investigation of the Genetic Variations and Candidate Genes Responsible for Seed Weight via GWAS in Paper Mulberry.Int J Mol Sci. 2022 Oct 19;23(20):12520. doi: 10.3390/ijms232012520. Int J Mol Sci. 2022. PMID: 36293375 Free PMC article.
-
OsEXPA7 Encoding an Expansin Affects Grain Size and Quality Traits in Rice (Oryza sativa L.).Rice (N Y). 2024 May 23;17(1):36. doi: 10.1186/s12284-024-00715-x. Rice (N Y). 2024. PMID: 38780864 Free PMC article.
-
CsJAZ11 positively regulates tea plant resistance to Colletotrichum camelliae via transient silencing and overexpression.Plant Cell Rep. 2025 Jul 13;44(8):174. doi: 10.1007/s00299-025-03563-1. Plant Cell Rep. 2025. PMID: 40652444
References
-
- Che, R. H. , Tong, H. N. , Shi, B. H. , Liu, Y. Q. , Fang, S. R. , Liu, D. P. , Xiao, Y. H. , Hu, B. , Liu, L. C. , Wang, H. R. , Zhao, M. F. , & Chu, C. C. (2016). Control of grain size and rice yield by GL2‐mediated brassinosteroid response. Nature Plants, 2, 15195–15201. 10.1038/nplants.2015.195 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

