MDIG, a 2‑oxoglutarate‑dependent oxygenase, acts as an oncogene and predicts the prognosis of multiple types of cancer
- PMID: 35583005
- PMCID: PMC9162052
- DOI: 10.3892/ijo.2022.5372
MDIG, a 2‑oxoglutarate‑dependent oxygenase, acts as an oncogene and predicts the prognosis of multiple types of cancer
Abstract
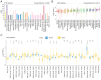
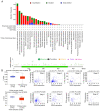
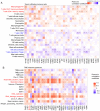
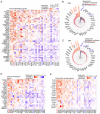
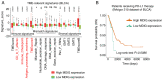
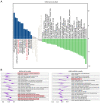
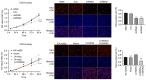
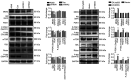
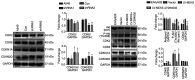
Recent studies have indicated that mineral dust‑induced gene (MDIG) is an oncogene induced by environmental factors, which has a key role in the development and progression of various tumor types, through epigenetic modifications; however, there are no previous pan‑cancer analyses of MDIG. In the present study, a comprehensive pan‑cancer analysis of MDIG was performed using public databases. The results demonstrated that MDIG was upregulated in tumor tissue samples compared with normal tissue, that it was present in all cancer cell lines and it was closely associated with the prognosis of patients with different tumor types. Furthermore, MDIG expression was closely associated with the immunological characteristics of the tumor microenvironment (TME), such as the frequency of tumor‑infiltrating immune cells, TME‑relevant signatures, immunostimulatory genes, immune checkpoint genes, chemokine receptor genes, tumor mutational burden and microsatellite instability. In parallel, high expression of MDIG was associated with improved overall survival of patients and this was verified in a cohort of patients who had received anti‑programmed cell death 1 ligand 1 treatment. Furthermore, high expression of MDIG led to multiple drug resistance in The Cancer Genome Atlas‑lung adenocarcinoma cohort. In addition, gene set variant analysis and gene set enrichment analysis indicated that MDIG was involved in cell cycle regulation. In vitro experiments suggested that MDIG promoted cell proliferation through the mTOR complex 2/Akt and pyruvate dehydrogenase kinase 1/Akt signaling pathways. In summary, the present study suggests that MDIG may be a prognostic biomarker and therapeutic target for various cancer types.
Keywords: AKT signaling; cell proliferation; mineral dust‑induced gene; pan‑cancer; prognostic and therapeutic biomarker.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



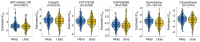
Similar articles
-
Pathological and Prognostic Indications of the mdig Gene in Human Lung Cancer.Cell Physiol Biochem. 2021 Jan 11;55(S2):13-28. doi: 10.33594/000000322. Cell Physiol Biochem. 2021. PMID: 33423409 Free PMC article.
-
The mineral dust-induced gene, mdig, regulates angiogenesis and lymphangiogenesis in lung adenocarcinoma by modulating the expression of VEGF-A/C/D via EGFR and HIF-1α signaling.Oncol Rep. 2021 May;45(5):60. doi: 10.3892/or.2021.8011. Epub 2021 Mar 24. Oncol Rep. 2021. PMID: 33760153
-
Mdig promotes oncogenic gene expression through antagonizing repressive histone methylation markers.Theranostics. 2020 Jan 1;10(2):602-614. doi: 10.7150/thno.36220. eCollection 2020. Theranostics. 2020. PMID: 31903140 Free PMC article.
-
New discoveries of mdig in the epigenetic regulation of cancers.Semin Cancer Biol. 2019 Aug;57:27-35. doi: 10.1016/j.semcancer.2019.06.013. Epub 2019 Jul 2. Semin Cancer Biol. 2019. PMID: 31276784 Free PMC article. Review.
-
Cooperation between NRF2-mediated transcription and MDIG-dependent epigenetic modifications in arsenic-induced carcinogenesis and cancer stem cells.Semin Cancer Biol. 2021 Nov;76:310-318. doi: 10.1016/j.semcancer.2021.03.030. Epub 2021 Apr 3. Semin Cancer Biol. 2021. PMID: 33823236 Free PMC article. Review.
Cited by
-
Epigenetics of Thymic Epithelial Tumors.Cancers (Basel). 2023 Jan 5;15(2):360. doi: 10.3390/cancers15020360. Cancers (Basel). 2023. PMID: 36672310 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
