Bayesian Analyses of Comparative Data with the Ornstein-Uhlenbeck Model: Potential Pitfalls
- PMID: 35583306
- PMCID: PMC9558839
- DOI: 10.1093/sysbio/syac036
Bayesian Analyses of Comparative Data with the Ornstein-Uhlenbeck Model: Potential Pitfalls
Abstract
The Ornstein-Uhlenbeck (OU) model is widely used in comparative phylogenetic analyses to study the evolution of quantitative traits. It has been applied to various purposes, including the estimation of the strength of selection or ancestral traits, inferring the existence of several selective regimes, or accounting for phylogenetic correlation in regression analyses. Most programs implementing statistical inference under the OU model have resorted to maximum-likelihood (ML) inference until the recent advent of Bayesian methods. A series of issues have been noted for ML inference using the OU model, including parameter nonidentifiability. How these problems translate to a Bayesian framework has not been studied much to date and is the focus of the present article. In particular, I aim to assess the impact of the choice of priors on parameter estimates. I show that complex interactions between parameters may cause the priors for virtually all parameters to impact inference in sometimes unexpected ways, whatever the purpose of inference. I specifically draw attention to the difficulty of setting the prior for the selection strength parameter, a task to be undertaken with much caution. I particularly address investigators who do not have precise prior information, by highlighting the fact that the effect of the prior for one parameter is often only visible through its impact on the estimate of another parameter. Finally, I propose a new parameterization of the OU model that can be helpful when prior information about the parameters is not available. [Bayesian inference; Brownian motion; Ornstein-Uhlenbeck model; phenotypic evolution; phylogenetic comparative methods; prior distribution; quantitative trait evolution.].
© The Author(s) 2022. Published by Oxford University Press, on behalf of the Society of Systematic Biologists.
Figures

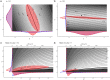
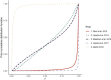
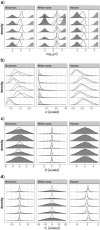
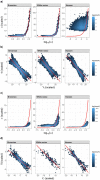

References
-
- Ané C. 2008. Analysis of comparative data with hierarchical autocorrelation. Ann. Appl. Stat. 2(3):1078–1102.
-
- Ané C., Ho L.S.T., Roch S.. 2017. Phase transition on the convergence rate of parameter estimation under an Ornstein-Uhlenbeck diffusion on a tree. J. Math. Biol. 74(1):355–385. - PubMed
-
- Beaulieu J.M., Jhwueng D.-C., Boettiger C., O’Meara B.C.. 2012. Modeling stabilizing selection: expanding the Ornstein-Uhlenbeck model of adaptive evolution. Evolution 66(8):2369–2383. - PubMed
-
- Blomberg S.P., Garland T.J., Ives A.R.. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57(4):717–745. - PubMed

