An induced pluripotent stem cell t(7;12)(q36;p13) acute myeloid leukemia model shows high expression of MNX1 and a block in differentiation of the erythroid and megakaryocytic lineages
- PMID: 35583991
- PMCID: PMC9545334
- DOI: 10.1002/ijc.34122
An induced pluripotent stem cell t(7;12)(q36;p13) acute myeloid leukemia model shows high expression of MNX1 and a block in differentiation of the erythroid and megakaryocytic lineages
Abstract
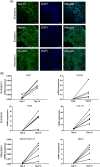
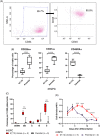
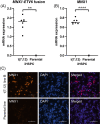
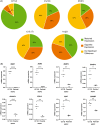
Acute myeloid leukemia (AML) results from aberrant hematopoietic processes and these changes are frequently initiated by chromosomal translocations. One particular subtype, AML with translocation t(7;12)(q36;p13), is found in children diagnosed before 2 years of age. The mechanisms for leukemogenesis induced by t(7;12) is not understood, in part because of the lack of efficient methods to reconstruct the leukemia-associated genetic aberration with correct genomic architecture and regulatory elements. We therefore created induced pluripotent stem cell (iPSC) lines that carry the translocation t(7;12) using CRISPR/Cas9. These t(7;12) iPSC showed propensity to differentiate into all three germ layers, confirming retained stem cell properties. The potential for differentiation into hematopoietic stem and progenitor cells (HSPC) was shown by expression of CD34, CD43 and CD45. Compared with the parental iPSC line, a significant decrease in cells expressing CD235a and CD41a was seen in the t(7;12) iPSC-derived HSPC (iHSPC), suggesting a block in differentiation. Moreover, colony formation assay showed an accumulation of cells at the erythroid and myeloid progenitor stages. Gene expression analysis revealed significant down-regulation of genes associated with megakaryocyte differentiation and up-regulation of genes associated with myeloid pathways but also genes typically seen in AML cases with t(7;12). Thus, this iPSC t(7;12) leukemia model of the t(7;12) AML subtype constitutes a valuable tool for further studies of the mechanisms for leukemia development and to find new treatment options.
Keywords: AML; chromosome translocation; leukemia; pluripotency; stem cells.
© 2022 The Authors. International Journal of Cancer published by John Wiley & Sons Ltd on behalf of UICC.
Conflict of interest statement
The authors declare no conflict of interest associated with this paper.
Figures


References
-
- Espersen ADL, Noren‐Nystrom U, Abrahamsson J, et al. Acute myeloid leukemia (AML) with t(7;12)(q36;p13) is associated with infancy and trisomy 19: data from Nordic Society for Pediatric Hematology and Oncology (NOPHO‐AML) and review of the literature. Genes Chromosomes Cancer. 2018;57:359‐365. - PubMed
-
- Tosi S, Hughes J, Scherer SW, et al. Heterogeneity of the 7q36 breakpoints in the t(7;12) involving ETV6 in infant leukemia. Genes Chromosomes Cancer. 2003;38:191‐200. - PubMed
-
- Ballabio E, Cantarella CD, Federico C, et al. Ectopic expression of the HLXB9 gene is associated with an altered nuclear position in t(7;12) leukemias. Leukemia. 2009;23:1179‐1182. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

