Diverse lineages of pathogenic Leptospira species are widespread in the environment in Puerto Rico, USA
- PMID: 35584143
- PMCID: PMC9154103
- DOI: 10.1371/journal.pntd.0009959
Diverse lineages of pathogenic Leptospira species are widespread in the environment in Puerto Rico, USA
Abstract
Background: Leptospirosis, caused by Leptospira bacteria, is a common zoonosis worldwide, especially in the tropics. Reservoir species and risk factors have been identified but surveys for environmental sources are rare. Furthermore, understanding of environmental Leptospira containing virulence associated genes and possibly capable of causing disease is incomplete, which may convolute leptospirosis diagnosis, prevention, and epidemiology.
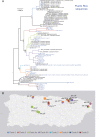
Methodology/principal findings: We collected environmental samples from 22 sites in Puerto Rico during three sampling periods over 14-months (Dec 2018-Feb 2020); 10 water and 10 soil samples were collected at each site. Samples were screened for DNA from potentially pathogenic Leptospira using the lipL32 PCR assay and positive samples were sequenced to assess genetic diversity. One urban site in San Juan was sampled three times over 14 months to assess persistence in soil; live leptospires were obtained during the last sampling period. Isolates were whole genome sequenced and LipL32 expression was assessed in vitro. We detected pathogenic Leptospira DNA at 15/22 sites; both soil and water were positive at 5/15 sites. We recovered lipL32 sequences from 83/86 positive samples (15/15 positive sites) and secY sequences from 32/86 (10/15 sites); multiple genotypes were identified at 12 sites. These sequences revealed significant diversity across samples, including four novel lipL32 phylogenetic clades within the pathogenic P1 group. Most samples from the serially sampled site were lipL32 positive at each time point. We sequenced the genomes of six saprophytic and two pathogenic Leptospira isolates; the latter represent a novel pathogenic Leptospira species likely belonging to a new serogroup.
Conclusions/significance: Diverse and novel pathogenic Leptospira are widespread in the environment in Puerto Rico. The disease potential of these lineages is unknown but several were consistently detected for >1 year in soil, which could contaminate water. This work increases understanding of environmental Leptospira diversity and should improve leptospirosis surveillance and diagnostics.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Vincent AT, Schiettekatte O, Goarant C, Neela VK, Bernet E, Thibeaux R, et al.. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl Trop Dis. 2019;13(5):e0007270. doi: 10.1371/journal.pntd.0007270 ; PubMed Central PMCID: PMC6532842. - DOI - PMC - PubMed
-
- Picardeau M, Bulach DM, Bouchier C, Zuerner RL, Zidane N, Wilson PJ, et al.. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One. 2008;3(2):e1607. doi: 10.1371/journal.pone.0001607 ; PubMed Central PMCID: PMC2229662. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

