Molecular and genomic investigation of an urban outbreak of dengue virus serotype 2 in Angola, 2017-2019
- PMID: 35584153
- PMCID: PMC9166355
- DOI: 10.1371/journal.pntd.0010255
Molecular and genomic investigation of an urban outbreak of dengue virus serotype 2 in Angola, 2017-2019
Abstract
Background: The transmission patterns and genetic diversity of dengue virus (DENV) circulating in Africa remain poorly understood. Circulation of the DENV serotype 1 (DENV1) in Angola was detected in 2013, while DENV serotype 2 (DENV2) was detected in 2018. Here, we report results from molecular and genomic investigations conducted at the Ministry of Health national reference laboratory (INIS) in Angola on suspected dengue cases detected between January 2017 and February 2019.
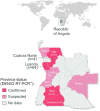
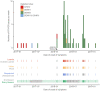
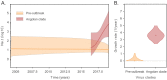
Methods: A total of 401 serum samples from dengue suspected cases were collected in 13 of the 18 provinces in Angola. Of those, 351 samples had complete data for demographic and epidemiological analysis, including age, gender, province, type of residence, clinical symptoms, as well as dates of onset of symptoms and sample collection. RNA was extracted from residual samples and tested for DENV-RNA using two distinct real time RT-PCR protocols. On-site whole genome nanopore sequencing was performed on RT-PCR+ samples. Bayesian coalescent models were used to estimate date and origin of outbreak emergence, as well as population growth rates.
Results: Molecular screening showed that 66 out of 351 (19%) suspected cases were DENV-RNA positive across 5 provinces in Angola. DENV RT-PCR+ cases were detected more frequently in urban sites compared to rural sites. Of the DENV RT-PCR+ cases most were collected within 6 days of symptom onset. 93% of infections were confirmed by serotype-specific RT-PCR as DENV2 and 1 case (1.4%) was confirmed as DENV1. Six CHIKV RT-PCR+ cases were also detected during the study period, including 1 co-infection of CHIKV with DENV1. Most cases (87%) were detected in Luanda during the rainy season between April and October. Symptoms associated with severe dengue were observed in 11 patients, including 2 with a fatal outcome. On-site nanopore genome sequencing followed by genetic analysis revealed an introduction of DENV2 Cosmopolitan genotype (also known as DENV2-II genotype) possibly from India in or around October 2015, at least 1 year before its detection in the country. Coalescent models suggest relatively moderately rapid epidemic growth rates and doubling times, and a moderate expansion of DENV2 in Angola during the studied period.
Conclusion: This study describes genomic, epidemiological and demographic characteristic of predominately urban transmission of DENV2 in Angola. We also find co-circulation of DENV2 with DENV1 and CHIKV and report several RT-PCR confirmed severe dengue cases in the country. Increasing dengue awareness in healthcare professional, expanding the monitorization of arboviral epidemics across the country, identifying most common mosquito breeding sites in urban settings, implementing innovative vector control interventions and dengue vaccination campaigns could help to reduce vector presence and DENV transmission in Angola.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- WHO. Dengue and severe dengue. World Health Organization; [Internet]. 2019.
-
- Gibbons RV, Kalanarooj S, Jarman RG, Nisalak A, Vaughn DW, Endy TP, et al.. Analysis of repeat hospital admissions for dengue to estimate the frequency of third or fourth dengue infections resulting in admissions and dengue hemorrhagic fever, and serotype sequences. The American journal of tropical medicine and hygiene. 2007;77(5):910–3. Epub 2007/11/07. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

