Ex silico engineering of cystine-dense peptides yielding a potent bispecific T cell engager
- PMID: 35584229
- PMCID: PMC10118748
- DOI: 10.1126/scitranslmed.abn0402
Ex silico engineering of cystine-dense peptides yielding a potent bispecific T cell engager
Abstract
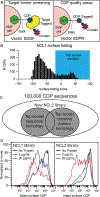
Cystine-dense peptides (CDPs) are a miniprotein class that can drug difficult targets with high affinity and low immunogenicity. Tools for their design, however, are not as developed as those for small-molecule and antibody drugs. CDPs have diverse taxonomic origins, but structural characterization is lacking. Here, we adapted Iterative Threading ASSEmbly Refinement (I-TASSER) and Rosetta protein modeling software for structural prediction of 4298 CDP scaffolds and performed in silico prescreening for CDP binders to targets of interest. Mammalian display screening of a library of docking-enriched, methionine and tyrosine scanned (DEMYS) CDPs against PD-L1 yielded binders from four distinct CDP scaffolds. One was affinity-matured, and cocrystallography yielded a high-affinity (KD = 202 pM) PD-L1-binding CDP that competes with PD-1 for PD-L1 binding. Its subsequent incorporation into a CD3-binding bispecific T cell engager produced a molecule with pM-range in vitro T cell killing potency and which substantially extends survival in two different xenograft tumor-bearing mouse models. Both in vitro and in vivo, the CDP-incorporating bispecific molecule outperformed a comparator antibody-based molecule. This CDP modeling and DEMYS technique can accelerate CDP therapeutic development.
Conflict of interest statement
Figures

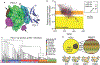





References
-
- Coleman N, Rodon J, Taking Aim at the Undruggable, Am. Soc. Clin. Oncol. Educ. B., e145–e152 (2021). - PubMed
-
- Busby RW, Kessler MM, Bartolini WP, Bryant AP, Hannig G, Higgins CS, Solinga RM, Tobin JV, Wakefield JD, Kurtz CB, Currie MG, Pharmacologic Properties, Metabolism, and Disposition of Linaclotide, a Novel Therapeutic Peptide Approved for the Treatment of Irritable Bowel Syndrome with Constipation and Chronic Idiopathic Constipation, J. Pharmacol. Exp. Ther. 344, 196–206 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

