Intrinsic transcriptional heterogeneity in neuroblastoma guides mechanistic and therapeutic insights
- PMID: 35584622
- PMCID: PMC9133465
- DOI: 10.1016/j.xcrm.2022.100632
Intrinsic transcriptional heterogeneity in neuroblastoma guides mechanistic and therapeutic insights
Abstract
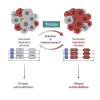
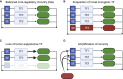
Cell state is controlled by master transcription factors (mTFs) that determine the cellular gene expression program. Cancer cells acquire dysregulated gene expression programs by mutational and non-mutational processes. Intratumoral heterogeneity can result from cells displaying distinct mTF-regulated cell states, which co-exist within the tumor. One archetypal tumor associated with transcriptionally regulated heterogeneity is high-risk neuroblastoma (NB). Patients with NB have poor overall survival despite intensive therapies, and relapsed patients are commonly refractory to treatment. The cellular populations that comprise NB are marked by different cohorts of mTFs and differential sensitivity to conventional therapies. Recent studies have highlighted mechanisms by which NB cells dynamically shift the cell state with treatment, revealing new opportunities to control the cellular response to treatment by manipulating cell-state-defining transcriptional programs. Here, we review recent advances in understanding transcriptionally defined cancer heterogeneity. We offer challenges to the field to encourage translation of basic science into clinical benefit.
Keywords: cell state; core regulatory circuitry; epigenetics; heterogeneity; neuroblastoma.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Turati V.A., Guerra-Assunção J.A., Potter N.E., Gupta R., Ecker S., Daneviciute A., Tarabichi M., Webster A.P., Ding C., May G., et al. Chemotherapy induces canalization of cell state in childhood B-cell precursor acute lymphoblastic leukemia. Nat. Cancer. 2021;2:835–852. doi: 10.1038/s43018-021-00219-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

