Broad and ultra-potent cross-clade neutralization of HIV-1 by a vaccine-induced CD4 binding site bovine antibody
- PMID: 35584627
- PMCID: PMC9133467
- DOI: 10.1016/j.xcrm.2022.100635
Broad and ultra-potent cross-clade neutralization of HIV-1 by a vaccine-induced CD4 binding site bovine antibody
Abstract
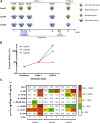
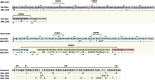
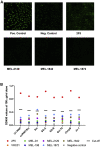
Human immunodeficiency virus type 1 (HIV-1) vaccination of cows has elicited broadly neutralizing antibodies (bNAbs). In this study, monoclonal antibodies (mAbs) are isolated from a clade A (KNH1144 and BG505) vaccinated cow using a heterologous clade B antigen (AD8). CD4 binding site (CD4bs) bNAb (MEL-1872) is more potent than a majority of CD4bs bNAbs isolated so far. MEL-1872 mAb with CDRH3 of 57 amino acids shows more potency (geometric mean half-maximal inhibitory concentration [IC50]: 0.009 μg/mL; breadth: 66%) than VRC01 against clade B viruses (29-fold) and than CHO1-31 against tested clade A viruses (21-fold). It also shows more breadth and potency than NC-Cow1, the only other reported anti-HIV-1 bovine bNAb, which has 60% breadth with geometric mean IC50 of 0.090 μg/mL in this study. Using successive different stable-structured SOSIP trimers in bovines can elicit bNAbs focusing on epitopes ubiquitous across subtypes. Furthermore, the cross-clade selection strategy also results in ultra-potent bNAbs.
Keywords: CDRH3; HIV-1; antibody; antiretroviral; cows; envelope glycoprotein; neutralization; vaccine.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests B.H. and D.F.J.P. are inventors on a corresponding patent from University of Melbourne: HIV-1 antibodies PCT/AU2021/050593. The other authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

