Distinct phenotypic states and spatial distribution of CD8+ T cell clonotypes in human brain metastases
- PMID: 35584630
- PMCID: PMC9133402
- DOI: 10.1016/j.xcrm.2022.100620
Distinct phenotypic states and spatial distribution of CD8+ T cell clonotypes in human brain metastases
Abstract
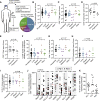
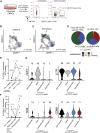
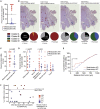
Metastatic disease in the brain is difficult to control and predicts poor prognosis. Here, we analyze human brain metastases and demonstrate their robust infiltration by CD8+ T cell subsets with distinct antigen specificities, phenotypic states, and spatial localization within the tumor microenvironment. Brain metastases are densely infiltrated by T cells; the majority of infiltrating CD8+ T cells express PD-1. Single-cell RNA sequencing shows significant clonal overlap between proliferating and exhausted CD8+ T cells, but these subsets have minimal clonal overlap with circulating and other tumor-infiltrating CD8+ T cells, including bystander CD8+ T cells specific for microbial antigens. Using spatial transcriptomics and spatial T cell receptor (TCR) sequencing, we show these clonally unrelated, phenotypically distinct CD8+ T cell populations occupy discrete niches within the brain metastasis tumor microenvironment. Together, our work identifies signaling pathways within CD8+ T cells and in their surrounding environment that may be targeted for immunotherapy of brain metastases.
Keywords: CD8(+) T cells; TCR-sequencing; brain metastases; bystander; exhaustion; spatial transcriptomics.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.A. holds patents related to the PD-1 pathway. All other authors declare no competing interests.
Figures





References
-
- Forrester J.V., McMenamin P.G., Dando S.J. CNS infection and immune privilege. Nat. Rev. Neurosci. 2018;19:655–671. - PubMed
-
- Berghoff A.S., Fuchs E., Ricken G., Mlecnik B., Bindea G., Spanberger T., Hackl M., Widhalm G., Dieckmann K., Prayer D., et al. Density of tumor-infiltrating lymphocytes correlates with extent of brain edema and overall survival time in patients with brain metastases. Oncoimmunology. 2016;5:e1057388. - PMC - PubMed
-
- Duchnowska R., Pęksa R., Radecka B., Mandat T., Trojanowski T., Jarosz B., Czartoryska-Arłukowicz B., Olszewski W.P., Och W., Kalinka-Warzocha E., et al. Immune response in breast cancer brain metastases and their microenvironment: the role of the PD-1/PD-L axis. Breast Cancer Res. 2016;18:43. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

