Large-scale GWAS of food liking reveals genetic determinants and genetic correlations with distinct neurophysiological traits
- PMID: 35585065
- PMCID: PMC9117208
- DOI: 10.1038/s41467-022-30187-w
Large-scale GWAS of food liking reveals genetic determinants and genetic correlations with distinct neurophysiological traits
Abstract
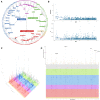
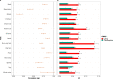
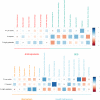
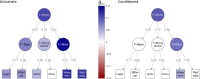
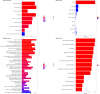
We present the results of a GWAS of food liking conducted on 161,625 participants from the UK-Biobank. Liking was assessed over 139 specific foods using a 9-point scale. Genetic correlations coupled with structural equation modelling identified a multi-level hierarchical map of food-liking with three main dimensions: "Highly-palatable", "Acquired" and "Low-caloric". The Highly-palatable dimension is genetically uncorrelated from the other two, suggesting that independent processes underlie liking high reward foods. This is confirmed by genetic correlations with MRI brain traits which show with distinct associations. Comparison with the corresponding food consumption traits shows a high genetic correlation, while liking exhibits twice the heritability. GWAS analysis identified 1,401 significant food-liking associations which showed substantial agreement in the direction of effects with 11 independent cohorts. In conclusion, we created a comprehensive map of the genetic determinants and associated neurophysiological factors of food-liking.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Costell E, Tárrega A, Bayarri S. Food acceptance: the role of consumer perception and attitudes. Chemosens. Percept. 2010;3:42–50. doi: 10.1007/s12078-009-9057-1. - DOI
-
- Hayes, J. E. In Handbook of Eating and Drinking 131–155 (Springer International Publishing, 2020).
Publication types
MeSH terms
Grants and funding
- RC2 MH089951/MH/NIMH NIH HHS/United States
- BHF_/British Heart Foundation/United Kingdom
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- MC_UU_00007/10/MRC_/Medical Research Council/United Kingdom
- MR/M016560/1/MRC_/Medical Research Council/United Kingdom
- R37 DA018673/DA/NIDA NIH HHS/United States
- BRC-1215-2001/DH_/Department of Health/United Kingdom
- U24 MH068457/MH/NIMH NIH HHS/United States
- 212904/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- R01 DA018673/DA/NIDA NIH HHS/United States
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- 102215/2/13/2/WT_/Wellcome Trust/United Kingdom
- 202802/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- G9815508/MRC_/Medical Research Council/United Kingdom
- 217065/Z/19/Z/WT_/Wellcome Trust/United Kingdom
- MC_PC_19009/MRC_/Medical Research Council/United Kingdom
- R01 MH081802/MH/NIMH NIH HHS/United States
- R01 DA042157/DA/NIDA NIH HHS/United States
- MC_UU_00011/1/MRC_/Medical Research Council/United Kingdom
- C18281/A29019/CRUK_/Cancer Research UK/United Kingdom
- EP-C-15-001/EPA/EPA/United States
- MC_PC_15018/MRC_/Medical Research Council/United Kingdom
- R01 DK092127/DK/NIDDK NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom

