Deubiquitinating enzymes and the proteasome regulate preferential sets of ubiquitin substrates
- PMID: 35585066
- PMCID: PMC9117253
- DOI: 10.1038/s41467-022-30376-7
Deubiquitinating enzymes and the proteasome regulate preferential sets of ubiquitin substrates
Abstract
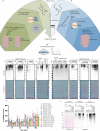
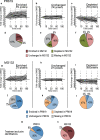
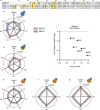
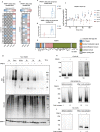
The ubiquitin-proteasome axis has been extensively explored at a system-wide level, but the impact of deubiquitinating enzymes (DUBs) on the ubiquitinome remains largely unknown. Here, we compare the contributions of the proteasome and DUBs on the global ubiquitinome, using UbiSite technology, inhibitors and mass spectrometry. We uncover large dynamic ubiquitin signalling networks with substrates and sites preferentially regulated by DUBs or by the proteasome, highlighting the role of DUBs in degradation-independent ubiquitination. DUBs regulate substrates via at least 40,000 unique sites. Regulated networks of ubiquitin substrates are involved in autophagy, apoptosis, genome integrity, telomere integrity, cell cycle progression, mitochondrial function, vesicle transport, signal transduction, transcription, pre-mRNA splicing and many other cellular processes. Moreover, we show that ubiquitin conjugated to SUMO2/3 forms a strong proteasomal degradation signal. Interestingly, PARP1 is hyper-ubiquitinated in response to DUB inhibition, which increases its enzymatic activity. Our study uncovers key regulatory roles of DUBs and provides a resource of endogenous ubiquitination sites to aid the analysis of substrate specific ubiquitin signalling.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests. UbiSite is patented by the University of Southern Denmark (patent number US9476888B2).
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

