PHGDH heterogeneity potentiates cancer cell dissemination and metastasis
- PMID: 35585241
- PMCID: PMC9888363
- DOI: 10.1038/s41586-022-04758-2
PHGDH heterogeneity potentiates cancer cell dissemination and metastasis
Erratum in
-
Author Correction: PHGDH heterogeneity potentiates cancer cell dissemination and metastasis.Nature. 2022 Sep;609(7927):E8. doi: 10.1038/s41586-022-05226-7. Nature. 2022. PMID: 36042336 Free PMC article. No abstract available.
Abstract
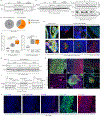
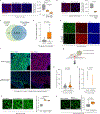
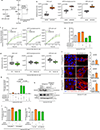
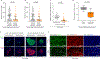
Cancer metastasis requires the transient activation of cellular programs enabling dissemination and seeding in distant organs1. Genetic, transcriptional and translational heterogeneity contributes to this dynamic process2,3. Metabolic heterogeneity has also been observed4, yet its role in cancer progression is less explored. Here we find that the loss of phosphoglycerate dehydrogenase (PHGDH) potentiates metastatic dissemination. Specifically, we find that heterogeneous or low PHGDH expression in primary tumours of patients with breast cancer is associated with decreased metastasis-free survival time. In mice, circulating tumour cells and early metastatic lesions are enriched with Phgdhlow cancer cells, and silencing Phgdh in primary tumours increases metastasis formation. Mechanistically, Phgdh interacts with the glycolytic enzyme phosphofructokinase, and the loss of this interaction activates the hexosamine-sialic acid pathway, which provides precursors for protein glycosylation. As a consequence, aberrant protein glycosylation occurs, including increased sialylation of integrin αvβ3, which potentiates cell migration and invasion. Inhibition of sialylation counteracts the metastatic ability of Phgdhlow cancer cells. In conclusion, although the catalytic activity of PHGDH supports cancer cell proliferation, low PHGDH protein expression non-catalytically potentiates cancer dissemination and metastasis formation. Thus, the presence of PHDGH heterogeneity in primary tumours could be considered a sign of tumour aggressiveness.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
DECLARATION OF INTERESTS
S-MF has received funding from Bayer AG, Merck and Black Belt Therapeutics, has consulted for Fund+ and is in the advisory board of Alesta Therapeutics. TGPG has consulted for Boehringer Ingelheim. MTL is an uncompensated President, CEO, and Limited Partner of StemMed Ltd. and an uncompensated Manager in StemMed Holdings LP, its General Partner. He is a founder and equity holder in Tvardi Therapeutics Inc., and a faculty member at Baylor College of Medicine. LED is a compensated employee at Baylor College of Medicine. All other authors declare no competing interests.
Figures










Comment in
-
Metabolic diversity drives cancer cell invasion.Nature. 2022 May;605(7911):627-628. doi: 10.1038/d41586-022-01301-1. Nature. 2022. PMID: 35585263 No abstract available.
References
REFERENCES MAIN TEXT
REFERENCES METHODS
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

