Evaluation of statistical approaches for association testing in noisy drug screening data
- PMID: 35585485
- PMCID: PMC9118710
- DOI: 10.1186/s12859-022-04693-z
Evaluation of statistical approaches for association testing in noisy drug screening data
Abstract
Background: Identifying associations among biological variables is a major challenge in modern quantitative biological research, particularly given the systemic and statistical noise endemic to biological systems. Drug sensitivity data has proven to be a particularly challenging field for identifying associations to inform patient treatment.
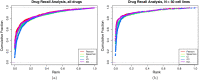
Results: To address this, we introduce two semi-parametric variations on the commonly used concordance index: the robust concordance index and the kernelized concordance index (rCI, kCI), which incorporate measurements about the noise distribution from the data. We demonstrate that common statistical tests applied to the concordance index and its variations fail to control for false positives, and introduce efficient implementations to compute p-values using adaptive permutation testing. We then evaluate the statistical power of these coefficients under simulation and compare with Pearson and Spearman correlation coefficients. Finally, we evaluate the various statistics in matching drugs across pharmacogenomic datasets.
Conclusions: We observe that the rCI and kCI are better powered than the concordance index in simulation and show some improvement on real data. Surprisingly, we observe that the Pearson correlation was the most robust to measurement noise among the different metrics.
Keywords: Association testing; Biomarker; Drug sensitivity; Non-parametric statistics; Pharmacogenomics; Power analysis; Statistics.
© 2022. The Author(s).
Conflict of interest statement
BHK is a shareholder and paid consultant for Code Ocean Inc. The authors have no other competing interests to declare.
Figures



References
MeSH terms
LinkOut - more resources
Full Text Sources

