Breast cancer risks associated with missense variants in breast cancer susceptibility genes
- PMID: 35585550
- PMCID: PMC9116026
- DOI: 10.1186/s13073-022-01052-8
Breast cancer risks associated with missense variants in breast cancer susceptibility genes
Abstract
Background: Protein truncating variants in ATM, BRCA1, BRCA2, CHEK2, and PALB2 are associated with increased breast cancer risk, but risks associated with missense variants in these genes are uncertain.
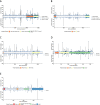
Methods: We analyzed data on 59,639 breast cancer cases and 53,165 controls from studies participating in the Breast Cancer Association Consortium BRIDGES project. We sampled training (80%) and validation (20%) sets to analyze rare missense variants in ATM (1146 training variants), BRCA1 (644), BRCA2 (1425), CHEK2 (325), and PALB2 (472). We evaluated breast cancer risks according to five in silico prediction-of-deleteriousness algorithms, functional protein domain, and frequency, using logistic regression models and also mixture models in which a subset of variants was assumed to be risk-associated.
Results: The most predictive in silico algorithms were Helix (BRCA1, BRCA2 and CHEK2) and CADD (ATM). Increased risks appeared restricted to functional protein domains for ATM (FAT and PIK domains) and BRCA1 (RING and BRCT domains). For ATM, BRCA1, and BRCA2, data were compatible with small subsets (approximately 7%, 2%, and 0.6%, respectively) of rare missense variants giving similar risk to those of protein truncating variants in the same gene. For CHEK2, data were more consistent with a large fraction (approximately 60%) of rare missense variants giving a lower risk (OR 1.75, 95% CI (1.47-2.08)) than CHEK2 protein truncating variants. There was little evidence for an association with risk for missense variants in PALB2. The best fitting models were well calibrated in the validation set.
Conclusions: These results will inform risk prediction models and the selection of candidate variants for functional assays and could contribute to the clinical reporting of gene panel testing for breast cancer susceptibility.
Keywords: Breast cancer; Genetic epidemiology; Missense variants; Risk prediction.
© 2022. The Author(s).
Conflict of interest statement
BV and SMH are employees and shareholders of Bio-Prodict, Nijmegen, The Netherlands. The remaining authors declare that they have no competing interests.
Figures

References
-
- Easton DF, Deffenbaugh AM, Pruss D, Frye C, Wenstrup RJ, Allen-Brady K, et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer–predisposition genes. Am J Human Gene. 2007;81(5):873–883. doi: 10.1086/521032. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

