Multiomics Analysis Identifies SOCS1 as Restraining T Cell Activation and Preventing Graft-Versus-Host Disease
- PMID: 35585676
- PMCID: PMC9313503
- DOI: 10.1002/advs.202200978
Multiomics Analysis Identifies SOCS1 as Restraining T Cell Activation and Preventing Graft-Versus-Host Disease
Abstract
Graft-versus-host disease (GVHD) is a major life-threatening complication of allogeneic hematopoietic stem cell transplantation (allo-HSCT). Inflammatory signaling pathways promote T-cell activation and are involved in the pathogenesis of GVHD. Suppressor of cytokine signaling 1 (SOCS1) is a critical negative regulator for several inflammatory cytokines. However, its regulatory role in T-cell activation and GVHD has not been elucidated. Multiomics analysis of the transcriptome and chromatin structure of granulocyte-colony-stimulating-factor (G-CSF)-administered hyporesponsive T cells from healthy donors reveal that G-CSF upregulates SOCS1 by reorganizing the chromatin structure around the SOCS1 locus. Parallel in vitro and in vivo analyses demonstrate that SOCS1 is critical for restraining T cell activation. Loss of Socs1 in T cells exacerbates GVHD pathogenesis and diminishes the protective role of G-CSF in GVHD mouse models. Further analysis shows that SOCS1 inhibits T cell activation not only by inhibiting the colony-stimulating-factor 3 receptor (CSF3R)/Janus kinase 2 (JAK2)/signal transducer and activator of transcription 3 (STAT3) pathway, but also by restraining activation of the inflammasome signaling pathway. Moreover, high expression of SOCS1 in T cells from patients correlates with low acute GVHD occurrence after HSCT. Overall, these findings identify that SOCS1 is critical for inhibiting T cell activation and represents a potential target for the attenuation of GVHD.
Keywords: SOCS1; T cell tolerance; graft-versus-host disease (GVHD); hematopoietic stem cell transplantation (HSCT); multiomics.
© 2022 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

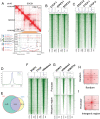
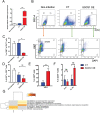



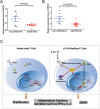
References
-
- a) Appelbaum F. R., N. Engl. J. Med. 2007, 357, 1472; - PubMed
- b) Dohner H., Estey E., Grimwade D., Amadori S., Appelbaum F. R., Buchner T., Dombret H., Ebert B. L., Fenaux P., Larson R. A., Levine R. L., Lo‐Coco F., Naoe T., Niederwieser D., Ossenkoppele G. J., Sanz M., Sierra J., Tallman M. S., Tien H. F., Wei A. H., Lowenberg B., Bloomfield C. D., Blood 2017, 129, 424; - PMC - PubMed
- c) Wang Y., Liu Q. F., Xu L. P., Liu K. Y., Zhang X. H., Ma X., Fan Z. P., Wu D. P., Huang X. J., Blood 2015, 125, 3956. - PubMed
-
- a) Markey K. A., MacDonald K. P., Hill G. R., Blood 2014, 124, 354; - PMC - PubMed
- b) Penack O., Marchetti M., Ruutu T., Aljurf M., Bacigalupo A., Bonifazi F., Ciceri F., Cornelissen J., Malladi R., Duarte R. F., Giebel S., Greinix H., Holler E., Lawitschka A., Mielke S., Mohty M., Arat M., Nagler A., Passweg J., Schoemans H., Socie G., Solano C., Vrhovac R., Zeiser R., Kroger N., Basak G. W., Lancet Haematol. 2020, 7, e157. - PubMed
-
- Pasquini MC Z. X., http://www.cibmtr.org (accessed: 2014).
-
- Zeiser R., Socie G., Blazar B. R., Br. J. Haematol. 2016, 175, 191. - PubMed
-
- a) Ferrara J. L., Reddy P., Semin. Hematol. 2006, 43, 3; - PubMed
- b) Burman A. C., Banovic T., Kuns R. D., Clouston A. D., Stanley A. C., Morris E. S., Rowe V., Bofinger H., Skoczylas R., Raffelt N., Fahy O., McColl S. R., Engwerda C. R., McDonald K. P., Hill G. R., Blood 2007, 110, 1064; - PubMed
- c) Tawara I., Koyama M., Liu C., Toubai T., Thomas D., Evers R., Chockley P., Nieves E., Sun Y., Lowler K. P., Malter C., Nishimoto N., Hill G. R., Reddy P., Clin. Cancer Res. 2011, 17, 77. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 2017YFA0104500/National Key Research and Development Program of China
- Z181110009618032/Beijing Municipal Science and Technology Commission
- 81670168/National Natural Science Foundation of China
- 81621001/National Natural Science Foundation of China
- 81530046/National Natural Science Foundation of China
- 81770189/National Natural Science Foundation of China
- Sino-Russian Mathematics Center
- 7204319/Beijing Municipal Natural Science Foundation
- 81230013/Key Program of the National Natural Science Foundation of China
- 2019-I2M-5-034/CAMS Innovation Fund for Medical Sciences
- Fundamental Research Funds for the Central Universities
- 31871266/National Natural Science Foundation of China
- 2021YFA1100300/National Key Research and Development Program of China
LinkOut - more resources
Full Text Sources
Miscellaneous
