Seroreactivity of the Severe Acute Respiratory Syndrome Coronavirus 2 Recombinant S Protein, Receptor-Binding Domain, and Its Receptor-Binding Motif in COVID-19 Patients and Their Cross-Reactivity With Pre-COVID-19 Samples From Malaria-Endemic Areas
- PMID: 35585976
- PMCID: PMC9109707
- DOI: 10.3389/fimmu.2022.856033
Seroreactivity of the Severe Acute Respiratory Syndrome Coronavirus 2 Recombinant S Protein, Receptor-Binding Domain, and Its Receptor-Binding Motif in COVID-19 Patients and Their Cross-Reactivity With Pre-COVID-19 Samples From Malaria-Endemic Areas
Abstract
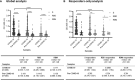
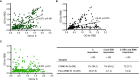
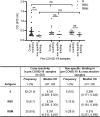
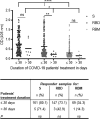
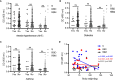
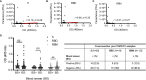
Despite the global interest and the unprecedented number of scientific studies triggered by the COVID-19 pandemic, few data are available from developing and low-income countries. In these regions, communities live under the threat of various transmissible diseases aside from COVID-19, including malaria. This study aims to determine the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) seroreactivity of antibodies from COVID-19 and pre-COVID-19 samples of individuals in Mali (West Africa). Blood samples from COVID-19 patients (n = 266) at Bamako Dermatology Hospital (HDB) and pre-COVID-19 donors (n = 283) from a previous malaria survey conducted in Dangassa village were tested by ELISA to assess IgG antibodies specific to the full-length spike (S) protein, the receptor-binding domain (RBD), and the receptor-binding motif (RBM436-507). Study participants were categorized by age, gender, treatment duration for COVID-19, and comorbidities. In addition, the cross-seroreactivity of samples from pre-COVID-19, malaria-positive patients against the three antigens was assessed. Recognition of the SARS-CoV-2 proteins by sera from COVID-19 patients was 80.5% for S, 71.1% for RBD, and 31.9% for RBM (p < 0.001). While antibody responses to S and RBD tended to be age-dependent, responses to RBM were not. Responses were not gender-dependent for any of the antigens. Higher antibody levels to S, RBD, and RBM at hospital entry were associated with shorter treatment durations, particularly for RBD (p < 0.01). In contrast, higher body weights negatively influenced the anti-S antibody response, and asthma and diabetes weakened the anti-RBM antibody responses. Although lower, a significant cross-reactive antibody response to S (21.9%), RBD (6.7%), and RBM (8.8%) was detected in the pre-COVID-19 and malaria samples. Cross-reactive antibody responses to RBM were mostly associated (p < 0.01) with the absence of current Plasmodium falciparum infection, warranting further study.
Keywords: COVID-19 samples; Pre-COVID-19 samples; SARS-CoV-2 S protein; cross-reactivity; malaria endemic-area; seroreactivity.
Copyright © 2022 Traoré, Guindo, Konaté, Traoré, Diakité, Kanté, Dembélé, Cissé, Incandela, Kodio, Coulibaly, Faye, Kajava, Pratesi, Migliorini, Papini, Pacini, Rovero, Errante, Diakité, Arevalo-Herrera, Herrera, Corradin and Balam.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

