Molecular Subtyping of Cancer Based on Distinguishing Co-Expression Modules and Machine Learning
- PMID: 35586568
- PMCID: PMC9108363
- DOI: 10.3389/fgene.2022.866005
Molecular Subtyping of Cancer Based on Distinguishing Co-Expression Modules and Machine Learning
Abstract
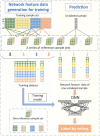
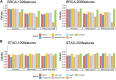
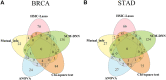
Molecular subtyping of cancer is recognized as a critical and challenging step towards individualized therapy. Most existing computational methods solve this problem via multi-classification of gene-expressions of cancer samples. Although these methods, especially deep learning, perform well in data classification, they usually require large amounts of data for model training and have limitations in interpretability. Besides, as cancer is a complex systemic disease, the phenotypic difference between cancer samples can hardly be fully understood by only analyzing single molecules, and differential expression-based molecular subtyping methods are reportedly not conserved. To address the above issues, we present here a new framework for molecular subtyping of cancer through identifying a robust specific co-expression module for each subtype of cancer, generating network features for each sample by perturbing correlation levels of specific edges, and then training a deep neural network for multi-class classification. When applied to breast cancer (BRCA) and stomach adenocarcinoma (STAD) molecular subtyping, it has superior classification performance over existing methods. In addition to improving classification performance, we consider the specific co-expressed modules selected for subtyping to be biologically meaningful, which potentially offers new insight for diagnostic biomarker design, mechanistic studies of cancer, and individualized treatment plan selection.
Keywords: machine learning; molecular subtyping of cancer; multi-classification; network perturbation; specific co-expression module.
Copyright © 2022 Sun, Wu, Yin, Jiang, Xu and Sun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources

