Refinements and considerations for trio whole-genome sequence analysis when investigating Mendelian diseases presenting in early childhood
- PMID: 35586607
- PMCID: PMC9108978
- DOI: 10.1016/j.xhgg.2022.100113
Refinements and considerations for trio whole-genome sequence analysis when investigating Mendelian diseases presenting in early childhood
Abstract
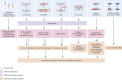
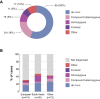
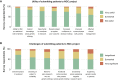
To facilitate early deployment of whole-genome sequencing (WGS) for severely ill children, a standardized pipeline for WGS analysis with timely turnaround and primary care pediatric uptake is needed. We developed a bioinformatics pipeline for comprehensive gene-agnostic trio WGS analysis of children suspected of having an undiagnosed monogenic disease that included detection and interpretation of primary genetic mechanisms of disease, including SNVs/indels, CNVs/SVs, uniparental disomy (UPD), imprinted genes, short tandem repeat expansions, mobile element insertions, SMN1/2 copy number calling, and mitochondrial genome variants. We assessed primary care practitioner experience and competence in a large cohort of 521 families (comprising 90% WGS trios). Children were identified by primary practitioners for recruitment, and we used the UK index of multiple deprivation to confirm lack of patient socio-economic status ascertainment bias. Of the 521 children sequenced, 176 (34%) received molecular diagnoses, with rates as high as 45% for neurology clinics. Twenty-three of the diagnosed cases (13%) required bespoke methods beyond routine SNV/CNV analysis. In our multidisciplinary clinician user experience assessment, both pediatricians and clinical geneticists expressed strong support for rapid WGS early in the care pathway, but requested further training in determining patient selection, consenting, and variant interpretation. Rapid trio WGS provides an efficacious single-pass screening test for children when deployed by primary practitioners in clinical settings that carry high a priori risk for rare pediatric disease presentations.
Keywords: genomics; mendelian disorders; paediatrics; rapid diagnostic whole genome; rare disease.
© 2022 University of Cambridge.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- French C.E., Delon I., Dolling H., Sanchis-Juan A., Shamardina O., Megy K., Abbs S., Austin T., Bowdin S., Branco R.G., et al. Next Generation Children Project Whole genome sequencing reveals that genetic conditions are frequent in intensively ill children. Intensive Care Med. 2019;45:627–636. doi: 10.1007/s00134-019-05552-x. - DOI - PMC - PubMed
-
- Gubbels C.S., VanNoy G.E., Madden J.A., Copenheaver D., Yang S., Wojcik M.H., Gold N.B., Genetti C.A., Stoler J., Parad R.B., et al. Prospective, phenotype-driven selection of critically ill neonates for rapid exome sequencing is associated with high diagnostic yield. Genet. Med. 2020;22:736–744. doi: 10.1038/s41436-019-0708-6. - DOI - PMC - PubMed
-
- Sanford E.F., Clark M.M., Farnaes L., Williams M.R., Perry J.C., Ingulli E.G., Sweeney N.M., Doshi A., Gold J.J., Briggs B., et al. Rapid whole genome sequencing has clinical utility in children in the PICU. Pediatr. Crit. Care Med. 2019;20:1007–1020. doi: 10.1097/PCC.0000000000002056. - DOI - PMC - PubMed
-
- Australian Genomics Health Alliance Acute Care F., Lunke S., Eggers S., Wilson M., Patel C., Barnett C.P., Pinner J., Sandaradura S.A., Buckley M.F., Krzesinski E.I., et al. Feasibility of ultra-rapid exome sequencing in critically ill infants and children with suspected monogenic conditions in the Australian public health care system. JAMA. 2020;323:2503–2511. doi: 10.1001/jama.2020.7671. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

