The Cellular and Developmental Roles of Cullins, Neddylation, and the COP9 Signalosome in Dictyostelium discoideum
- PMID: 35586714
- PMCID: PMC9108976
- DOI: 10.3389/fphys.2022.827435
The Cellular and Developmental Roles of Cullins, Neddylation, and the COP9 Signalosome in Dictyostelium discoideum
Abstract
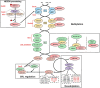
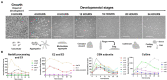
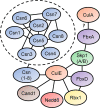
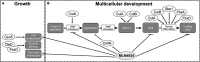
Cullins (CULs) are a core component of cullin-RING E3 ubiquitin ligases (CRLs), which regulate the degradation, function, and subcellular trafficking of proteins. CULs are post-translationally regulated through neddylation, a process that conjugates the ubiquitin-like modifier protein neural precursor cell expressed developmentally downregulated protein 8 (NEDD8) to target cullins, as well as non-cullin proteins. Counteracting neddylation is the deneddylase, COP9 signalosome (CSN), which removes NEDD8 from target proteins. Recent comparative genomics studies revealed that CRLs and the CSN are highly conserved in Amoebozoa. A well-studied representative of Amoebozoa, the social amoeba Dictyostelium discoideum, has been used for close to 100 years as a model organism for studying conserved cellular and developmental processes owing to its unique life cycle comprised of unicellular and multicellular phases. The organism is also recognized as an exceptional model system for studying cellular processes impacted by human diseases, including but not limited to, cancer and neurodegeneration. Recent work shows that the neddylation inhibitor, MLN4924 (Pevonedistat), inhibits growth and multicellular development in D. discoideum, which supports previous work that revealed the cullin interactome in D. discoideum and the roles of cullins and the CSN in regulating cellular and developmental processes during the D. discoideum life cycle. Here, we review the roles of cullins, neddylation, and the CSN in D. discoideum to guide future work on using this biomedical model system to further explore the evolutionarily conserved functions of cullins and neddylation.
Keywords: COP9 signalosome; Dictyostelium discoideum; SCF complex; cullins; neddylation.
Copyright © 2022 Kim, Mathavarajah and Huber.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Inhibiting Neddylation with MLN4924 Suppresses Growth and Delays Multicellular Development in Dictyostelium discoideum.Biomolecules. 2021 Mar 23;11(3):482. doi: 10.3390/biom11030482. Biomolecules. 2021. PMID: 33807046 Free PMC article.
-
Differential dynamics of cullin deneddylation via COP9 signalosome subunit 5 interaction.Biochem Biophys Res Commun. 2022 Dec 31;637:341-347. doi: 10.1016/j.bbrc.2022.11.045. Epub 2022 Nov 16. Biochem Biophys Res Commun. 2022. PMID: 36423380
-
Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation.Nat Cell Biol. 2005 Oct;7(10):1014-20. doi: 10.1038/ncb1301. Epub 2005 Aug 28. Nat Cell Biol. 2005. PMID: 16127432
-
Protection of cullin-RING E3 ligases by CSN-UBP12.Trends Cell Biol. 2006 Jul;16(7):362-9. doi: 10.1016/j.tcb.2006.05.001. Epub 2006 Jun 9. Trends Cell Biol. 2006. PMID: 16762551 Review.
-
Targeting the COP9 signalosome for cancer therapy.Cancer Biol Med. 2022 Mar 21;19(5):573-90. doi: 10.20892/j.issn.2095-3941.2021.0605. Cancer Biol Med. 2022. PMID: 35315259 Free PMC article. Review.
Cited by
-
Tracking of Ubiquitin Signaling through 3.5 Billion Years of Combinatorial Conjugation.Int J Mol Sci. 2024 Aug 8;25(16):8671. doi: 10.3390/ijms25168671. Int J Mol Sci. 2024. PMID: 39201358 Free PMC article. Review.
-
Naegleria genus pangenome reveals new structural and functional insights into the versatility of these free-living amoebae.Front Microbiol. 2023 Feb 1;13:1056418. doi: 10.3389/fmicb.2022.1056418. eCollection 2022. Front Microbiol. 2023. PMID: 36817109 Free PMC article.
-
Crosstalk between protein post-translational modifications and phase separation.Cell Commun Signal. 2024 Feb 12;22(1):110. doi: 10.1186/s12964-023-01380-1. Cell Commun Signal. 2024. PMID: 38347544 Free PMC article. Review.
-
Synergy between a cytoplasmic vWFA/VIT protein and a WD40-repeat F-box protein controls development in Dictyostelium.Front Cell Dev Biol. 2023 Sep 14;11:1259844. doi: 10.3389/fcell.2023.1259844. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37779900 Free PMC article.
-
Cullin-Conciliated Regulation of Plant Immune Responses: Implications for Sustainable Crop Protection.Plants (Basel). 2024 Oct 26;13(21):2997. doi: 10.3390/plants13212997. Plants (Basel). 2024. PMID: 39519916 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

