Host-Population Microbial Diversity Scaling of Chinese Gut Microbiomes in Gout Patients
- PMID: 35586773
- PMCID: PMC9109170
- DOI: 10.1177/11769343221095858
Host-Population Microbial Diversity Scaling of Chinese Gut Microbiomes in Gout Patients
Abstract
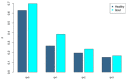
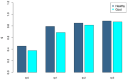
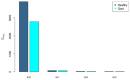
Gout is a prevalent chronic inflammatory disease that affects the life of tens of millions of people worldwide, and it typically presents as gout arthritis, gout stone, or even kidney damage. Research has revealed its connection with the gut microbiome, although exact mechanism is still unclear. Studies have shown the decline of microbiome diversity in gout patients and change of microbiome compositions between the gout patients and healthy controls. Nevertheless, how diversity changes across host individuals at a cohort (population) level has not been investigated to the best of our knowledge. Here we apply the diversity-area relationship (DAR), which is an extension to the classic SAR (species-area relationship) and establishes the power-function model between microbiome diversity and the number of individuals within cohort, to comparatively investigate diversity scaling (changes) of gut microbiome in gout patients and healthy controls. The DAR modeling with a study involving 83 subjects (41 gout patients) revealed that the potential number of microbial species in gout patients is only 70% of that in the healthy control (2790 vs 3900) although the difference may not be statistically significant. The other DAR parameters including diversity scaling and similarity parameters did not show statistically significant differences. We postulate that the high resilience of gut microbiome may explain the lack of significant gout-disease effects on gut microbial diversity at the population level. The lack of statistically significant difference between the gout patients and healthy controls at host population (cohort) level is different from the previous findings at individual level in the existing literature.
Keywords: Gout; diversity scaling parameter; diversity-area relationship (DAR); diversity-disease relationship; gut microbiome.
© The Author(s) 2022.
Conflict of interest statement
Declaration of Conflicting Interests: The author declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures
References
-
- Dalbeth N, Choi HK, Joosten LAB, et al. Gout. Nat Rev Dis Primers. 2019;5:69. - PubMed
-
- Richette P, Clerson P, Périssin L, Flipo RM, Bardin T. Revisiting comorbidities in gout: a cluster analysis. Ann Rheum Dis. 2015;74:142-147. - PubMed
-
- Smith E, Hoy D, Cross M, et al. The global burden of gout: estimates from the global burden of disease 2010 study. Ann Rheum Dis. 2014;73:1470-1476. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

