Visualization of mRNA Expression in Pseudomonas aeruginosa Aggregates Reveals Spatial Patterns of Fermentative and Denitrifying Metabolism
- PMID: 35586988
- PMCID: PMC9195945
- DOI: 10.1128/aem.00439-22
Visualization of mRNA Expression in Pseudomonas aeruginosa Aggregates Reveals Spatial Patterns of Fermentative and Denitrifying Metabolism
Abstract
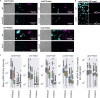
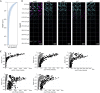
Gaining insight into the behavior of bacteria at the single-cell level is important given that heterogeneous microenvironments strongly influence microbial physiology. The hybridization chain reaction (HCR) is a technique that provides in situ molecular signal amplification, enabling simultaneous mapping of multiple target RNAs at small spatial scales. To refine this method for biofilm applications, we designed and validated new probes to visualize the expression of key catabolic genes in Pseudomonas aeruginosa aggregates. In addition to using existing probes for the dissimilatory nitrate reductase (narG), we developed probes for a terminal oxidase (ccoN1), nitrite reductase (nirS), nitrous oxide reductase (nosZ), and acetate kinase (ackA). These probes can be used to determine gene expression levels across heterogeneous populations such as biofilms. Using these probes, we quantified gene expression across oxygen gradients in aggregate populations grown using the
Keywords: ABBA; HCR; Pseudomonas aeruginosa; aggregates; biofilms; denitrification; gene expression; heterogeneity; hybridization chain reaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Quantitative Visualization of Gene Expression in Mucoid and Nonmucoid Pseudomonas aeruginosa Aggregates Reveals Localized Peak Expression of Alginate in the Hypoxic Zone.mBio. 2019 Dec 17;10(6):e02622-19. doi: 10.1128/mBio.02622-19. mBio. 2019. PMID: 31848278 Free PMC article.
-
Protein Network of the Pseudomonas aeruginosa Denitrification Apparatus.J Bacteriol. 2016 Apr 14;198(9):1401-13. doi: 10.1128/JB.00055-16. Print 2016 May. J Bacteriol. 2016. PMID: 26903416 Free PMC article.
-
Protein complex formation during denitrification by Pseudomonas aeruginosa.Microb Biotechnol. 2017 Nov;10(6):1523-1534. doi: 10.1111/1751-7915.12851. Epub 2017 Aug 31. Microb Biotechnol. 2017. PMID: 28857512 Free PMC article. Review.
-
Kinetics of nirS expression (cytochrome cd1 nitrite reductase) in Pseudomonas stutzeri during the transition from aerobic respiration to denitrification: evidence for a denitrification-specific nitrate- and nitrite-responsive regulatory system.J Bacteriol. 1999 Jan;181(1):161-6. doi: 10.1128/JB.181.1.161-166.1999. J Bacteriol. 1999. PMID: 9864326 Free PMC article.
-
Microenvironmental characteristics and physiology of biofilms in chronic infections of CF patients are strongly affected by the host immune response.APMIS. 2017 Apr;125(4):276-288. doi: 10.1111/apm.12668. APMIS. 2017. PMID: 28407427 Review.
Cited by
-
Targeting Anaerobic Respiration in Pseudomonas aeruginosa with Chlorate Improves Healing of Chronic Wounds.Adv Wound Care (New Rochelle). 2024 Feb;13(2):53-69. doi: 10.1089/wound.2023.0036. Epub 2023 Aug 14. Adv Wound Care (New Rochelle). 2024. PMID: 37432895 Free PMC article.
-
Spatial transcriptome-guided multi-scale framework connects P. aeruginosa metabolic states to oxidative stress biofilm microenvironment.PLoS Comput Biol. 2024 Apr 26;20(4):e1012031. doi: 10.1371/journal.pcbi.1012031. eCollection 2024 Apr. PLoS Comput Biol. 2024. PMID: 38669236 Free PMC article.
-
How to study biofilms: technological advancements in clinical biofilm research.Front Cell Infect Microbiol. 2023 Dec 13;13:1335389. doi: 10.3389/fcimb.2023.1335389. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38156318 Free PMC article. Review.
-
Mechanisms of microbial co-aggregation in mixed anaerobic cultures.Appl Microbiol Biotechnol. 2024 Jul 4;108(1):407. doi: 10.1007/s00253-024-13246-8. Appl Microbiol Biotechnol. 2024. PMID: 38963458 Free PMC article. Review.
-
Biofilm antimicrobial susceptibility through an experimental evolutionary lens.NPJ Biofilms Microbiomes. 2022 Oct 18;8(1):82. doi: 10.1038/s41522-022-00346-4. NPJ Biofilms Microbiomes. 2022. PMID: 36257971 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

