In-depth proteomics characterization of ∆Np73 effectors identifies key proteins with diagnostic potential implicated in lymphangiogenesis, vasculogenesis and metastasis in colorectal cancer
- PMID: 35586989
- PMCID: PMC9298678
- DOI: 10.1002/1878-0261.13228
In-depth proteomics characterization of ∆Np73 effectors identifies key proteins with diagnostic potential implicated in lymphangiogenesis, vasculogenesis and metastasis in colorectal cancer
Abstract
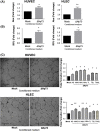
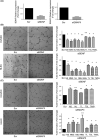
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer-related death worldwide. Alterations in proteins of the p53-family are a common event in CRC. ΔNp73, a p53-family member, shows oncogenic properties and its effectors are largely unknown. We performed an in-depth proteomics characterization of transcriptional control by ∆Np73 of the secretome of human colon cancer cells and validated its clinical potential. The secretome was analyzed using high-density antibody microarrays and stable isotopic metabolic labeling. Validation was performed by semiquantitative PCR, ELISA, dot-blot and western blot analysis. Evaluation of selected effectors was carried out using 60 plasma samples from CRC patients, individuals carrying premalignant colorectal lesions and colonoscopy-negative controls. In total, 51 dysregulated proteins were observed showing at least 1.5-foldchange in expression. We found an important association between the overexpression of ∆Np73 and effectors related to lymphangiogenesis, vasculogenesis and metastasis, such as brain-derived neurotrophic factor (BDNF) and the putative aminoacyl tRNA synthase complex-interacting multifunctional protein 1 (EMAP-II)-vascular endothelial growth factor C-vascular endothelial growth factor receptor 3 axis. We further demonstrated the usefulness of BDNF as a potential CRC biomarker able to discriminate between CRC patients and premalignant individuals from controls with high sensitivity and specificity.
Keywords: colorectal cancer; in-depth proteomics; lymphangiogenesis; secretome; ∆Np73 effectors.
© 2022 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Rodriguez‐Salas N, Dominguez G, Barderas R, Mendiola M, Garcia‐Albeniz X, Maurel J, et al. Clinical relevance of colorectal cancer molecular subtypes. Crit Rev Oncol Hematol. 2017;109:9–19. - PubMed
-
- Rodriguez N, Pelaez A, Barderas R, Dominguez G. Clinical implications of the deregulated TP73 isoforms expression in cancer. Clin Transl Oncol. 2018;20:827–36. - PubMed
-
- Irwin MS, Kaelin WG. p53 family update: p73 and p63 develop their own identities. Cell Growth Differ. 2001;12(7):337–49. - PubMed
-
- Jost CA, Marin MC, Kaelin WG Jr. p73 is a simian [correction of human] p53‐related protein that can induce apoptosis. Nature. 1997;389(6647):191–4. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

