Repertoires of SARS-CoV-2 epitopes targeted by antibodies vary according to severity of COVID-19
- PMID: 35587156
- PMCID: PMC9122311
- DOI: 10.1080/21505594.2022.2073025
Repertoires of SARS-CoV-2 epitopes targeted by antibodies vary according to severity of COVID-19
Abstract
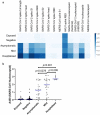
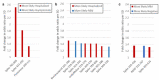
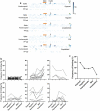
Antibodies to SARS-CoV-2 are central to recovery and immunity from COVID-19. However, the relationship between disease severity and the repertoire of antibodies against specific SARS-CoV-2 epitopes an individual develops following exposure remains incompletely understood. Here, we studied seroprevalence of antibodies to specific SARS-CoV-2 and other betacoronavirus antigens in a well-annotated, community sample of convalescent and never-infected individuals obtained in August 2020. One hundred and twenty-four participants were classified into five groups: previously exposed but without evidence of infection, having no known exposure or evidence of infection, seroconverted without symptoms, previously diagnosed with symptomatic COVID-19, and recovered after hospitalization with COVID-19. Prevalence of IgGs specific to the following antigens was compared between the five groups: recombinant SARS-CoV-2 and betacoronavirus spike and nucleocapsid protein domains, peptides from a tiled array of 22-mers corresponding to the entire spike and nucleocapsid proteins, and peptides corresponding to predicted immunogenic regions from other proteins of SARS-CoV-2. Antibody abundance generally correlated positively with severity of prior illness. A number of specific immunogenic peptides and some that may be associated with milder illness or protection from symptomatic infection were identified. No convincing association was observed between antibodies to Receptor Binding Domain(s) (RBDs) of less pathogenic betacoronaviruses HKU1 or OC43 and COVID-19 severity. However, apparent cross-reaction with SARS-CoV RBD was evident and some predominantly asymptomatic individuals had antibodies to both MERS-CoV and SARS-CoV RBDs. Findings from this pilot study may inform development of diagnostics, vaccines, and therapeutic antibodies, and provide insight into viral pathogenic mechanisms.
Keywords: SARS-CoV-2; antibodies; asymptomatic infection; epitopes; peptide array; seroprevalence.
Conflict of interest statement
T.E.M. discloses financial interest in Telomere Diagnostics and Care Access Research, unrelated to this manuscript. S.M., M.D.S., R.K.W., and R.D.P. are employees of Cell Signaling Technology, Inc. The other authors report no conflict of interest.
Figures



References
-
- Khoury DS, Cromer D, Reynaldi A, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27(7):1205–1211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
