Dietary wheat and reduced methane yield are linked to rumen microbiome changes in dairy cows
- PMID: 35587477
- PMCID: PMC9119556
- DOI: 10.1371/journal.pone.0268157
Dietary wheat and reduced methane yield are linked to rumen microbiome changes in dairy cows
Abstract
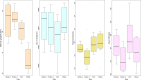
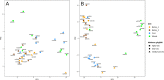
Fermentation of pasture grasses and grains in the rumen of dairy cows and other ruminants produces methane as a by-product, wasting energy and contributing to the atmospheric load of greenhouse gasses. Many feeding trials in farmed ruminants have tested the impact of dietary components on feed efficiency, productivity and methane yield (MeY). Such diets remodel the rumen microbiome, altering bacterial, archaeal, fungal and protozoan populations, with an altered fermentation outcome. In dairy cows, some dietary grains can reduce enteric methane production. This is especially true of wheat, in comparison to corn or barley. Using a feeding trial of cows fed rolled wheat, corn or barley grain, in combination with hay and canola, we identified wheat-associated changes in the ruminal microbiome. Ruminal methane production, pH and VFA concentration data together with 16S rRNA gene amplicon sequences were used to compare ruminal bacterial and archaeal populations across diets. Differential abundance analysis of clustered sequences (OTU) identified members of the bacterial families Lachnospiraceae, Acidaminococcaceae, Eubacteriaceae, Prevotellaceae, Selenomonadaceae, Anaerovoracaceae and Fibrobacteraceae having a strong preference for growth in wheat-fed cows. Within the methanogenic archaea, (at >99% 16S rRNA sequence identity) the growth of Methanobrevibacter millerae was favoured by the non-wheat diets, while Methanobrevibacter olleyae was unaffected. From the wheat-preferring bacteria, correlation analysis found OTU strongly linked to reduced MeY, reduced pH and raised propionic acid levels. OTU from the genera Shuttleworthia and Prevotella_7 and especially Selenomonadaceae had high anti-methane correlations. An OTU likely representing (100% sequence identity) the fumarate-reducing, hydrogen-utilising, rumen bacterium Mitsuokella jalaludinii, had an especially high negative correlation coefficient (-0.83) versus MeY and moderate correlation (-0.6) with rumen pH, strongly suggesting much of the MeY suppression is due to reduced hydrogen availablity. Other OTU, representing as yet unknown species from the Selenomonadaceae family and the genera Prevotella_7, Fibrobacter and Syntrophococcus also had high to moderate negative MeY correlations, but low correlation with pH. These latter likely represent bacterial species able to reduce MeY without causing greater ruminal acidity, making them excellent candidates, provided they can be isolated, for development as anti-methane probiotics.
Conflict of interest statement
No authors have competing interests.
Figures



References
-
- FP OM. The significance of livestock as a contributor to global greenhouse gas emissions today and in the near future. Fuel and Energy Abstracts. 2011;166(1):7–15.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

